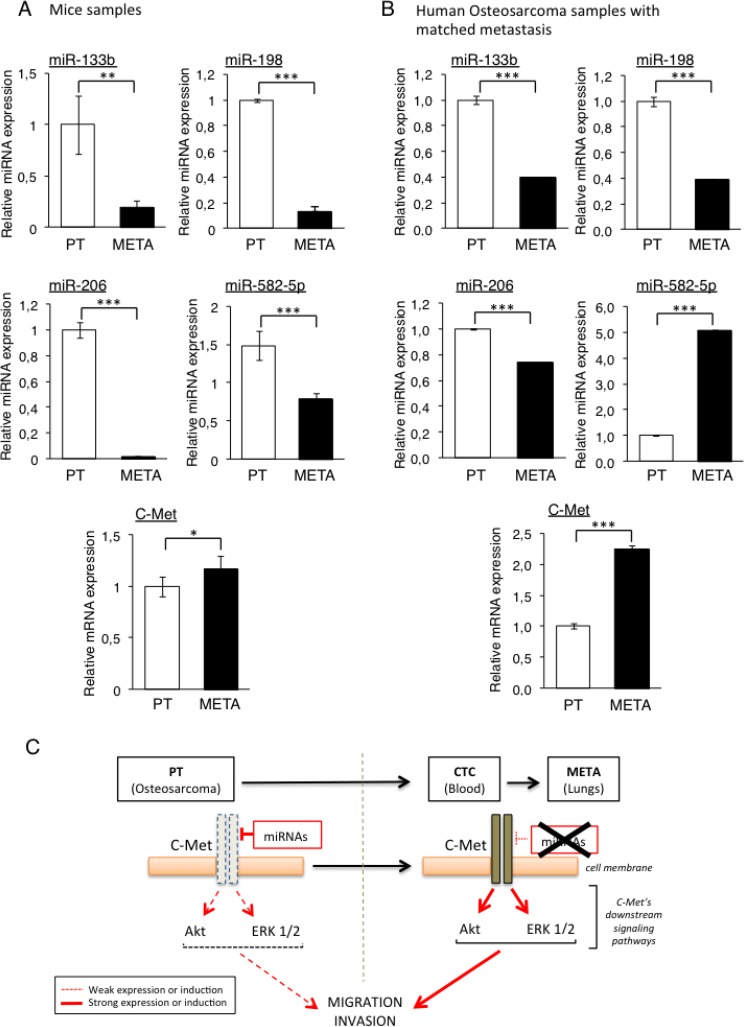
Figure 2. Validation of candidate miRNAs repression and C-Met overexpression in the lung-metastasis (METAs) samples from both mice and patients’ samples.
The four candidate miRNAs’ expression (miR-133b, -198, -206 and -582-5p) as well as C-Met were analysed by RT-qPCR from xenografted-Osteosarcoma from mice samples (A) and from four metastatic Osteosarcoma patients (B). The expression levels were compared between primary tumors (PTs) and their associated metastases (METs). *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA and Dunnett’s multiple comparison tests were used to compare the significance of the results. Average expression and error bars (s.d.) are represented for n = 3 measurements from representative experiments. RNU6B, GAPDH and B2M are used as housekeeping genes. (C) Rationale of the hypothetic mechanism: cells from the primary bone tumor (PT) express the candidates miRNAs predicted to target C-Met, thus maintaining a low activity of this receptor and its downstream signaling pathways, keeping the PT cells at their initial site. The miRNAs’ loss of expression in Circulating Tumor Cells (CTCs) and metastatic nodules (METAs) releases the pressure exerted on C-Met, resulting in increasing both its expression and activity, thus enhancing the metastatic potential of those cells.