Figure 6. Comparison of mapping depth using adaptive evolution and the deep scanning mutagenesis library.
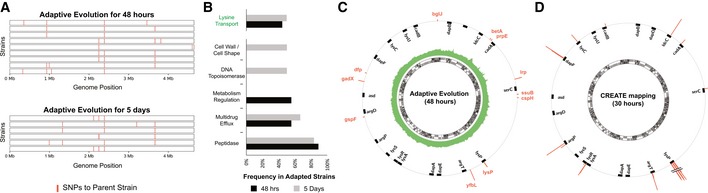
- Map of SNPs positions observed in each sequenced genome after 48‐h adaptation (n = 9) and 5‐day adaptation with one passage per day (n = 6). SNPs were mapped to the parent strain, sequenced after growth in minimal media in the absence of AEC.
- Categories of the SNPs found in Fig 6A, with the categories from genes that are directly linked to the lysine pathway highlighted in green.
- Circos plot of the SNPs found after 48‐hour adaptation relative to the lysine metabolism genes. The bar plots of each SNP represent the frequency across sequenced strains (n = 9). Lysine metabolism genes are highlighted as black bars (50× zoom). The inner green plot represents the average sequencing coverage for each position across all sequenced genomes.
- Map of enriched mutations found for the same selective pressure (1,000 μM AEC) using our deep scanning mutagenesis library. The bar plots represent log2 enrichment scores.
