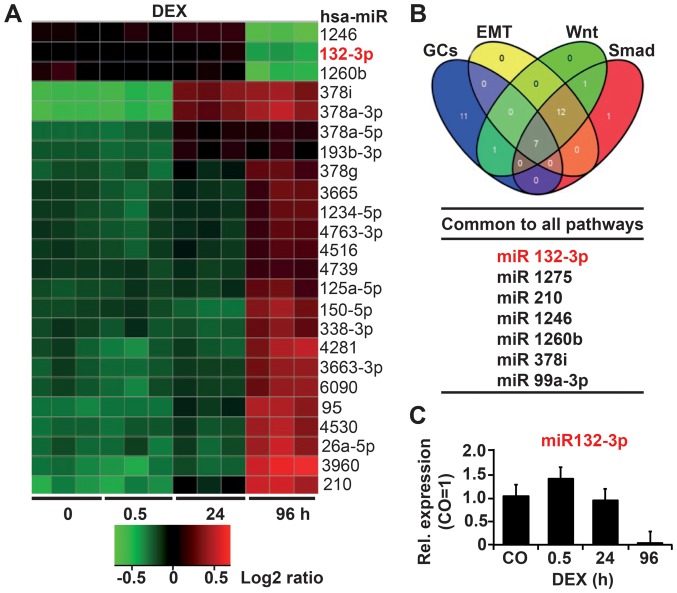
Figure 1.
DEX downregulates miR-132. (A) AsPC-1 cells were treated with 1 µM DEX or left as CO. Total RNA was extracted at 30 min, 24 and 96 h and analyzed by Agilent miRNA Microarray (Release 19.0). The heatmap includes the top 24 significantly differentially upregulated and downregulated miRNAs. The red color indicates high expression, and the green color indicates low expression. (B) A Venn diagram was created based on the top three candidates and the results of in silico analysis with the keywords glucocorticoids, EMT, Wnt and Smad using different bioinformatics algorithms. (C) AsPC-1 cells were treated with DEX as indicated or left untreated as a CO, followed by the isolation of total RNA and RT-qPCR analysis with primers specific for miR-132-3p. Expression levels were normalized to that of RNU6B. The mean fold change in the CO cells was set to 1; the expression levels in treated cells were relative to that of the CO. DEX, dexamethasone; miRNA, microRNA; EMT, epithelial mesenchymal transition; CO, control/untreated.