Significance
Drug-resistant subpopulations of microbes or tumor cells are difficult to detect but can confound disease treatment. In this deep characterization of a chronic fungal infection, we report unprecedented heterogeneity in the drug resistance-related gene MRR1 among Clavispora (Candida) lusitaniae isolates from a single individual. Cells expressing Mrr1 variants that led to drug resistance, by elevated expression of the MDR1-encoded efflux protein, were present at low levels in each sample and thus were undetected in standard assays. We provide evidence that these drug-resistant fungi may arise indirectly in response to other factors present in the infection. Our work suggests that alternative methods may be able to identify drug-resistant subpopulations and thus positively impact patient care.
Keywords: Candida, drug resistance, evolution, fungi, heterogeneity
Abstract
Management of the limited number of antimicrobials currently available requires the identification of infections that contain drug-resistant isolates and the discovery of factors that promote the evolution of drug resistance. Here, we report a single fungal infection in which we have identified numerous subpopulations that differ in their alleles of a single gene that impacts drug resistance. The diversity at this locus was markedly greater than the reported heterogeneity of alleles conferring antibiotic resistance in bacterial infections. Analysis of genomes from hundreds of Clavispora (Candida) lusitaniae isolates, through individual and pooled isolate sequencing, from a single individual with cystic fibrosis revealed at least 25 nonsynonymous mutations in MRR1, which encodes a transcription factor capable of inducing fluconazole (FLZ) resistance in Candida species. Isolates with high-activity Mrr1 variants were resistant to FLZ due to elevated expression of the MDR1-encoded efflux pump. We found that high Mrr1-regulated Mdr1 activity protected against host and bacterial factors, suggesting drug resistance can be selected for indirectly and perhaps explaining the Mrr1 heterogeneity in this individual who had no prior azole exposure. Regional analysis of C. lusitaniae populations from the upper and lower lobes of the right lung suggested intermingling of subpopulations throughout. Our retrospective characterization of sputum and lung populations by pooled sequencing found that alleles that confer FLZ resistance were a minority in each pool, possibly explaining why they were undetected before unsuccessful FLZ therapy. New susceptibility testing regimes may detect problematical drug-resistant subpopulations in heterogeneous single-species infections.
The limited number of available antimicrobials necessitates strategies to better enable their judicious use in appropriate cases to prevent further development of drug resistance (1). The ability to determine drug susceptibility in single-species infections can be complicated by heterogeneity within the infecting population. Heterogeneity in drug resistance can result from coinfections by phylogenetically distinct strains with allelic differences that affect drug sensitivities (2, 3). However, the diversification of microbes within chronic infections may be an even more important driver of allelic heterogeneity and the development of drug resistance differences. Analysis of Helicobacter pylori isolates from ulcers or Mycobacterium tuberculosis isolates from the lung have shown that isolates derived from the same strain can have different levels of antimicrobial resistance (4, 5). Diversification of bacteria within chronic lung infections associated with the genetic disease cystic fibrosis (CF) has also been shown to lead to heterogeneous drug resistances within the population (6–8).
The discovery of a CF patient with a high-burden chronic fungal infection containing phenotypically heterogeneous isolates provided the opportunity to analyze fungal population structure. These analyses led us to discover a striking, and possibly unprecedented, level of heterogeneity in the sequence of a single drug resistance-related gene among the haploid Clavispora (Candida) lusitaniae isolates that are otherwise genomically similar. Unlike some of the diploid Candida species, such as Candida albicans, which are common members of the human microbiome, C. lusitaniae is most often isolated from environmental samples. Like other Candida species, C. lusitaniae can cause both acute and long-term infections (9). C. lusitaniae is particularly notorious for its rapid development of resistance to multiple antifungals during therapy (10–12) and is phylogenetically closely related to Candida auris (13), multidrug-resistant strains of which have caused outbreaks in recent years (14).
Our work shows the presence of a complex, dynamic, and structured population of C. lusitaniae within a single infection. Through the analysis of over 300 C. lusitaniae isolates from a single patient we observed heterogeneity in fluconazole (FLZ) resistance and found that this heterogeneity was largely caused by the presence of at least 12 different alleles of MRR1, which encodes a drug-resistance regulator. The enrichment of nonsynonymous mutations in MRR1 greatly exceeded the heterogeneity at any other locus. Here we have explored factors that may have contributed to the selection for drug-resistant subpopulations and highlight the importance of assessing and treating fungal populations during the management of disease.
Results
C. lusitaniae Coisolates Are Phenotypically and Genotypically Heterogeneous.
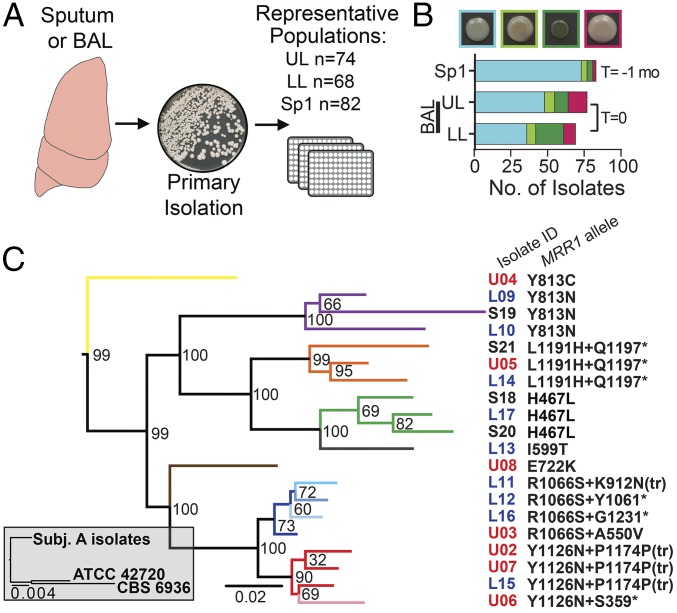
Analysis of bronchoalveolar lavage (BAL) fluid from a subject with CF detected >105 cfu/mL of C. lusitaniae in both the right upper lobe (UL) and right lower lobe (LL) of the lung with very few coinfecting bacteria (subject 6 in ref. 15, subject A here). C. lusitaniae, while rarely encountered in CF, has been detected in CF respiratory sputum previously (16). The patient history revealed non-albicans Candida species were detected in sputum cultures collected 6 mo before the BAL, suggesting that C. lusitaniae may have been present for a prolonged period. C. lusitaniae isolates recovered from UL and LL BAL fluid (n = 74 and 68 isolates, respectively; Fig. 1A) varied in colony color when grown on the chromogenic medium CHROMagar Candida (17, 18), indicating isolates differed in enzymatic activities (Fig. 1B). Additional C. lusitaniae isolates were obtained from an archived sputum sample (Sp1, n = 82 isolates) collected 1 mo before the BAL, confirming the persistence of C. lusitaniae in the lung for at least 1 mo. The Sp1 coisolates were also phenotypically heterogeneous on CHROMagar Candida medium (Fig. 1B).
Fig. 1.
C. lusitaniae coisolates are phenotypically and genomically heterogeneous. (A) Schematic of isolate acquisition from patient samples. Primary BAL fluid or sputum samples were plated. Numerous colonies from each plate were streak-purified and saved to represent the population within each sample. Following species identification by ITS1 sequencing, phenotypic and genomic analyses were performed for the indicated number of isolates per sample. (B) Phenotypic distribution of C. lusitaniae isolate colony color on CHROMagar Candida medium, example of colony colors shown above graph. From left to right, colors include blue/green, brown/green, green, and purple/mauve. The Sp1 (sputum) sample was obtained approximately 1 mo prior (T = −1 mo) to the UL and LL BAL samples (T = 0). (C) Gray inset contains a maximum-likelihood tree for the 20 subject A clinical isolates compared with ATCC 42720 and CBS 6936. The expanded maximum-likelihood tree shows the relationship between subject A isolates based on interisolate SNPs found through WGS, and bootstrap values are shown at every branch point; arms are colored by MRR1 allele. Isolate identifiers are color-coded by sample of origin: Sp1 (black), UL BAL (red), and LL BAL (blue). MRR1 alleles are denoted by the amino acid changes caused by nonsynonymous SNPs and INDELs; asterisk indicates stop codon. Amino acid numbers are based on the MRR1 reannotation in SI Appendix, Fig. S4. One nucleotide INDELs in codons 1174 and 912 cause an amino acid change in the latter case and frame shift mutations that resulted in premature stop codons, noted by “tr” for truncation, at N1176 and L927, respectively.
We performed whole-genome sequencing (WGS) for 20 isolates from the UL, LL, and Sp1 samples (n = 7, 9, and 4 isolates, respectively) chosen to represent different CHROMagar phenotypes. Genomic analysis indicated that the isolates were more closely related to each other than to other sequenced strains, ATCC 42720 and CBS 6936, and thus shared a recent common ancestor (Fig. 1C). Pairwise analyses of the 20 clinical isolate genomes found 24–131 SNPs between any two isolates, with 404 high-confidence interisolate SNPs (45% nonsynonymous) in total (SI Appendix, Fig. S1 and Dataset S1). Similarly, 76–179 insertions or deletions (INDELs) differed between any two isolates, with 536 INDELs in total. The INDELs were primarily short, with >60% being less than 3 nt in length and 80% being intergenic (SI Appendix, Fig. S1 and Dataset S2). Phylogenetic analysis using either SNPs or INDELs found similar relationships between isolates (Fig. 1C and SI Appendix, Fig. S2). Isolates did not cluster by the lavage or sputum sample of origin, indicating that genomic heterogeneity was not solely explained by spatial separation (UL vs. LL) or population changes over time (Sp1 vs. BAL). Additionally, phylogenetic analysis of SNPs and INDELs did not cluster isolates by colony color on CHROMagar Candida medium, suggesting this is likely a complex trait which we will not explore further here.
Although it is possible that some genomic heterogeneity was present within the original infecting inoculum, analysis of copy number variation has suggested that the genomes of these isolates were changing within the context of this infection (SI Appendix, Fig. S3A). We identified a duplication of chromosome 6R in the four Sp1 isolates obtained at the earlier time point, which suggests it may have been ancestral, that was repeatedly lost in UL and LL isolates from separate clusters within the cladogram (SI Appendix, Fig. S3B). Overall, however, the number of differences between the isolates is much smaller than that which is reported for differences between fungal strains within a species (19, 20) (Fig. 1C). Thus, these data indicate that these coisolates are recently diverged and are evolving within the infection in ways that lead to phenotypic heterogeneity.
Allelic Heterogeneity in MRR1 Confers Differences in FLZ Resistance.
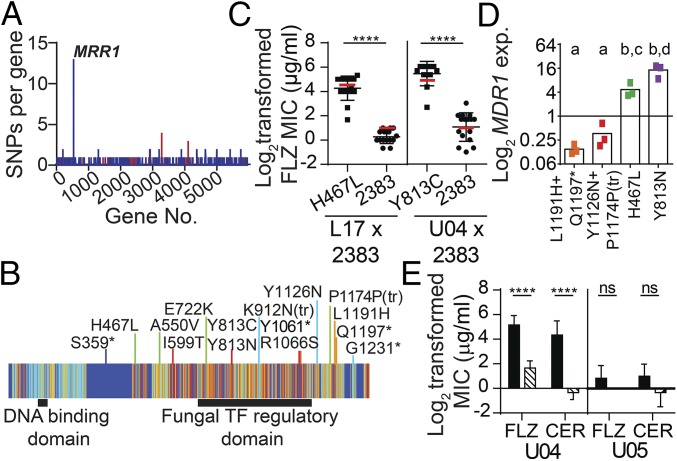
Evaluation of the number of mutations per gene found that the most heterogeneous locus among the 20 sequenced isolates was CLUG_00542, an ortholog of C. albicans MRR1 (SI Appendix, Fig. S4). CLUG_00542 is referred to hereafter as MRR1. Among the 20 C. lusitaniae clinical isolate genomes we found that MRR1 contained 13 nonsynonymous SNPs and two INDELs, but no synonymous SNPs, within 12 distinct MRR1 alleles (Figs. 1C and 2 A and B). No other gene contained more than two nonsynonymous SNPs and only a few genes contained more than two INDELs (Fig. 2A and Datasets S1 and S2). Phylogenetic analysis suggested that eight of the MRR1 alleles arose independently and the remaining four alleles had two mutations (two SNPs or one SNP and one INDEL) that arose sequentially (Fig. 1C). Alignment of C. lusitaniae Mrr1 with Mrr1 orthologs in other Candida species revealed that the SNPs and INDELs generally fell within regions of moderate to high protein sequence conservation (Fig. 2B). The absence of mutations in MRR1-adjacent genes indicated that this locus was not in a hypervariable region of the genome, and the absence of synonymous SNPs in MRR1 indicated that the sequence heterogeneity was not solely due to high rates of nucleotide substitution (Datasets S1 and S2). Together, these data suggest that MRR1 was under strong selection.
Fig. 2.
Multiple nonsynonymous SNPs in MRR1 increase FLZ resistance via up-regulation of MDR1 expression. (A) Number of nonsynonymous (blue) and synonymous (red) SNPs within each gene. (B) Schematic of Mrr1 depicting the locations of the amino acid changes caused by the 13 nonsynonymous SNPs and two INDELs. Mrr1 is represented by a heat map of sequence conservation, described in SI Appendix, Fig. S4, with increased conservation represented by a gradient from cool (dark blue) to warm (red) colors. The color of the line marking the location of each mutation corresponds to the sequence conservation score of the affected amino acid. (C) Log2-transformed FLZ MICs (micrograms per milliliter) of mating progeny, measured at 48 h, obtained by crossing the FLZS 2383 (MRR12383) strain to FLZR clinical isolate L17 (chxR, MRR1H467L, progeny n = 30) or U04 (chxR, MRR1Y813C, progeny n = 28); isolates are grouped by MRR1 allele. Red lines indicate the mean FLZ MIC for the parental strain for each MRR1 allele. Mean ± SD of three independent measurements are shown, ****P < 0.0001. (D) MDR1 expression (exp) for FLZS [MRR1L1191H+Q1197* and MRR1Y1126N+P1174P(tr)] and FLZR (MRR1H467L and MRR1Y813N) isolates with the same MRR1 allele (n = 3, colored to match phylogenetic tree in Fig. 1C). MDR1 expression was normalized to ACT1 levels. Data represent the average of three independent replicates, a-b and c-d P < 0.01. (E) FLZ and cerulenin (CER) MICs for WT (solid) and the isogenic mdr1Δ derivatives (striped) of the FLZR U04 (MRR1Y813C) and FLZS U05 (MRR1L1191H+Q1197*) isolates measured at 48 h. Mean ± SD for four independent replicates shown, ****P < 0.0001; ns, not significant.
Heterogeneity in MRR1 is of interest because prior studies in other Candida species have shown that constitutively active variants of the Mrr1 transcription factor can be selected for during azole therapy and are capable of conferring FLZ resistance (21–23). In this case, there was no history of antifungal treatment, yet a fraction of the 224 BAL and Sp1 isolates exhibited high resistance to FLZ, but not to other classes of antifungals including polyenes and echinocandins (amphotericin B and caspofungin, respectively; SI Appendix, Fig. S5). Further analysis revealed that isolates with the MRR1H467L (S18, S20, and L17) and MRR1Y813N (S19, L09, and L10) alleles had FLZ minimum inhibitory concentrations (MICs) of 16 μg/mL, which is clinically resistant and notably higher than the reported MIC50 for C. lusitaniae (0.5 μg/mL) (24). In contrast, isolates with the MRR1L1191H+Q1197* (S21, L14, and U05) and MRR1Y1126N+P1174P(tr) (L15, U02, and U07) alleles had FLZ MICs of 0.5–2 μg/mL (SI Appendix, Fig. S6A). Mating FLZ-resistant (FLZR) clinical isolates with an FLZ-sensitive (FLZS) strain of a distinct genetic background confirmed that Mrr1H467L and Mrr1Y813C variants conferred a high FLZ MIC (Fig. 2C; P < 0.0001). No FLZR progeny were detected in a similar mating assay using an FLZS clinical isolate (SI Appendix, Fig. S7). Thus, the heterogeneity in MRR1 alleles created a C. lusitaniae population with heterogeneous levels of resistance to FLZ.
Other genes known to impact FLZ resistance in Candida species like UPC2 (CLUG_03901) (25) and TAC1 (CLUG_02369) (26) did not contain SNPs or INDELs (Datasets S1 and S2). There was also no correlation between the chromosome 6R duplication, found in nine isolates, and FLZ MIC (SI Appendix, Fig. S3C). One isolate, U02 [MRR1Y1126N+P1174P(tr)], had a duplication of the left arm of chromosome 6 where ERG11 (CLUG_04932) is located (SI Appendix, Fig. S3). Erg11, a lanosterol 14α-demethylase, is a known target of FLZ and increased expression of ERG11 has been shown to increase FLZ resistance in other Candida species (27). Thus, this duplication may explain the slightly elevated FLZ MIC of U02 [MRR1Y1126N+P1174P(tr)] relative to other isolates with the same MRR1 allele (U07 and L15) (SI Appendix, Fig. S6A).
To assess the impact of differences in Mrr1 on the transcriptome, we performed an RNA sequencing (RNA-Seq) analysis and identified 19 differentially expressed genes between representative FLZR isolates, with Mrr1 variants Y813C (U04), Y813N (L10), or H467L (L17), and FLZS isolates, with Mrr1 variants L1191H+Q1197* (U05) or Y1126N+P1174P(tr) (U07) (SI Appendix, Table S1). One of the most differentially expressed genes was MDR1 [CLUG_01938/01939 (SI Appendix, Fig. S8) or MFS7 (28)], which encodes an ortholog of the C. albicans Mdr1 multidrug efflux transporter. In C. albicans, Mdr1 contributes to the elevated FLZ MIC in strains with constitutively active Mrr1 variants (22, 29, 30). Quantitative RT-PCR analysis found that MDR1 expression was similar among isolates, with the same MRR1 allele and again significantly higher in FLZR isolates than in FLZS isolates (Fig. 2D; P < 0.01). FLZ MIC correlated with MDR1 levels across all isolates (SI Appendix, Fig. S6C). MDR1 was deleted from isolates with either high Mrr1 activity (U04, MRR1Y813C) or low Mrr1 activity (U05, MRR1L1191H+Q1197*). Deletion of MDR1 in U04 caused a sixfold decrease in FLZ MIC and a 26-fold decrease in the MIC for cerulenin, another substrate of C. albicans Mdr1 (31). In contrast, deletion of MDR1 in U05 did not alter the MIC for these compounds (Fig. 2E). These data indicate that Mdr1 activity differs between isolates and can contribute to FLZR in C. lusitaniae.
In addition to MDR1, the C. lusitaniae Mrr1 regulon contained CLUG_02968 (Caorf19.7306), CLUG_01281, and CLUG_04991 (putative methylglyoxal reductases with sequence similarity to CaGRP2) and multiple putative oxidoreductases (SI Appendix, Table S1). Some of the C. albicans orthologs and genes with similar predicted functions are also regulated by C. albicans Mrr1 (30). MRR1 itself was not differentially expressed between the resistant and susceptible isolates.
High Mrr1 Activity Confers Resistance to Host and Microbial Factors.
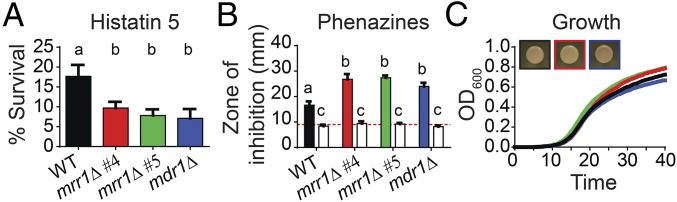
Although MRR1 appeared to be under positive selection within the C. lusitaniae population in the lung, even in the absence of antifungal treatment, this gene did not appear to be under strong selection in the laboratory (SI Appendix, Fig. S9). Passaging strains with either high (L17, MRR1H467L) or low (U05, MRR1L1191H+Q1197*) FLZ resistance in a defined medium did not yield populations with significantly different FLZ resistance profiles. Thus, we proposed that Mrr1 variants that confer high Mdr1 activity could have been selected for in response to immune system components or by factors produced by coinfecting microbes. Prior data have indicated that high Mdr1 activity in C. albicans contributes to resistance to histatin 5 (Hst 5), a peptide secreted by the salivary glands as part of the innate immune system (32, 33). We found that deletion of either MRR1 or MDR1 from the FLZR U04 (MRR1Y813C) isolate reduced survival in the presence Hst 5 by twofold (Fig. 3A; P < 0.001), confirming the necessity of Mrr1 and Mdr1 for Hst 5 resistance. In addition to host defenses, the lungs of patients with CF are typically polymicrobial environments filled with molecules produced by a variety of bacteria (15, 34). Phenazines, produced by the common CF pathogen Pseudomonas aeruginosa, are known to inhibit the growth and metabolism of some Candida species (35) and can be found at high concentrations in CF sputum (36). We found a role for high Mrr1 activity and concomitant high MDR1 expression in protection against these phenazine toxins. Deletion of either MRR1 or MDR1 from the FLZR isolate U04 (MRR1Y813C) increased the zone of clearance around phenazine-producing P. aeruginosa colonies (Fig. 3B; P < 0.0001). The U04 clinical isolate and its mrr1Δ and mdr1Δ derivatives grew equally well in the presence of P. aeruginosa colonies that could not produce phenazines (Fig. 3B). Analysis of all 20 clinical isolates revealed an inverse correlation between zone of inhibition due to phenazines and either FLZ MIC or MDR1 expression (SI Appendix, Fig. S6 D and E), suggesting Mrr1 activity correlates with phenazine resistance. Deletion of either MRR1 or MDR1 does not alter other phenotypes including growth rate or colony color on CHROMagar medium (Fig. 3C). While we cannot know if either Hst 5 or phenazines selected for high Mrr1 activity in these C. lusitaniae isolates, these examples demonstrate how strains with high Mrr1 activity might be more fit in vivo even in the absence of azole drugs.
Fig. 3.
High Mrr1 activity confers resistance to host and microbial factors. (A and B) The FLZR clinical isolate U04 (WT, MRR1Y813C, black) is more resistant than its isogenic mrr1Δ (red and green) and mdr1Δ (blue) derivatives to Hst 5 and phenazines. (A) Percent survival after a 1-h incubation with 3.75 μM Hst 5. Mean ± SD shown for three independent measurements, a-b P < 0.001. (B) Size of the zone of clearance or inhibition (millimeters) around colonies of P. aeruginosa strain PA14 WT (solid) or Δphz (open) on the indicated C. lusitaniae lawns. Red dotted line indicates the average size of the P. aeruginosa colonies. Mean ± SD for representative data shown, similar trends were observed in three independent replicates, a-b P < 0.0001, a-c and b-c P < 0.0001. (C) Growth curve and colony color on CHROMagar Candida medium of the U04 (WT, MRR1Y813C, black) isolate and its isogenic mrr1Δ (4, red and 5, green) and mdr1Δ (blue) derivatives. Representative data are shown, similar results in three independent replicates.
Isolates Containing Constitutively Active Mrr1 Variants Are Minor Members of the Population.
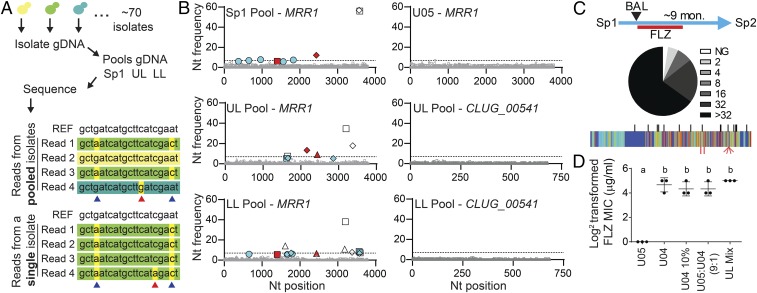
We performed an unbiased pooled sequencing analysis of ∼70 isolates from the UL, LL, and Sp1 samples (Fig. 1A). Single-isolate WGS data were used to establish thresholds for the identification of biallelic positions within the pooled WGS data (7% of reads per position for novel SNPs and 5% for previously confirmed SNPs) (Fig. 4A). In the Pool-Seq data, there was striking heterogeneity in nucleotide frequency within the MRR1 locus relative to single-isolate WGS data (Fig. 4B). The MRR1-adjacent gene CLUG_00541 and other loci analyzed were not similarly heterogeneous (Fig. 4B). Using the 5% threshold for previously confirmed SNPs, we found 11 of the 13 SNPs identified by single-isolate WGS within the pools. Of these, the MRR1 SNPs found in FLZR isolates (encoding H467L, E722K, Y813N, and Y813C Mrr1 variants) represented 18, 22, and 12% of the reads in the Sp1, UL, and LL pools, respectively (Fig. 4B, red symbols). Although the respective ratios of each SNP changed between samples, SNPs indicative of FLZR isolates were present at low levels in all three samples, even in the absence of prior antifungal therapy. Using a more stringent threshold for the detection of novel SNPs, those not found among the 20 sequenced genomes, we identified four new SNPs in MRR1, all of which were nonsynonymous, further underscoring the level of heterogeneity at this locus (Fig. 4B).
Fig. 4.
Small subpopulations of FLZR isolates can alter the FLZ resistance profile of the population. (A) Schematic for the creation of pooled isolate DNA. Equivalent amounts of DNA were obtained from individual isolates then combined before sequencing. Individual genomes were not uniquely marked. A representation of Illumina reads from pooled and single isolate sequencing is shown in comparison with the reference sequence (REF) from ATCC 42720. Differences in read color represent sequences that originated from different isolates; nucleotides highlighted in yellow differ from the reference. SNPs (blue arrows) are largely invariant (common across >95% of reads) in single-isolate sequence data but present at a lower frequency in pooled sequencing data. We determined the threshold for SNP detection in pools based on the rate of sequencing errors (red arrows) in single-isolate WGS data. (B) Nucleotide frequency of nonreference alleles for MRR1 from pooled (Sp1, UL, and LL) and single isolates (U05) WGS and CLUG_00541 from pooled (UL and LL) WGS. The frequencies of the nonreference nucleotides are plotted at every position (reference set to zero); positions that differ from the reference but are shared among the clinical isolates are not shown due to scale. Nucleotides are represented as follows: A (square), C (triangle), G (circle), and T (diamond). Nucleotide frequencies shown in gray are present at less than 5%, the threshold at which we are unable to distinguish between low-abundance alleles and sequencing error. Open symbols represent previously identified SNPs that correlate with FLZ MIC ≤ 8 μg/mL, red filled symbols represent previously identified SNPs that correlate with a FLZ MIC ≥ 16 μg/mL, and blue filled symbols represent SNPs that were not detected in any of the 20 single-isolate genomes. The dashed line at 7% represents the threshold for novel SNP detection; only blue nucleotides above 7% were counted as novel SNPs, of which there were four. (C) Lowest inhibitory concentration of FLZ for each Sp2 (n = 83) isolate, from no growth, NG, to >32 μg/mL FLZ (Sp1 data shown in SI Appendix, Fig. S5A) and schematic showing sample acquisition timeline including 4 mo of FLZ treatment and 5 mo off antifungals. Novel SNPs in MRR1 identified in Sp2 isolates (red, Bottom) are plotted on the sequence conservation heat map from Fig. 2, with SNPs and INDEL locations identified in Sp1, UL, and LL isolates marked in black (Top) for reference. (D) Log2-transformed FLZ MICs (micrograms per milliliter) measured at 24 h for U05 and U04 alone, U04 alone at 10% the normal starting concentration (U04 10%), a 9:1 mixture of U05:U04, and a mixture of equivalent amounts of all 74 UL isolates. Mean ± SD from three independent replicates is shown, a-b ****P < 0.0001.
Unrecognized FLZ-Resistant Subpopulations May Contribute to Treatment Failure.
To complete the longitudinal analysis of this infection, we analyzed an additional sputum sample from this subject (Sp2) collected after a 4-mo course of oral FLZ treatment (started after the BAL sample was collected) and 5 mo with no prescribed antifungal therapy. Abundant C. lusitaniae were found at this timepoint, indicating either FLZ treatment failed to clear this infection or that recolonization occurred after FLZ therapy was completed. Among the 83 Sp2 isolates analyzed, we found a range of FLZ susceptibilities (2 to >32 μg/mL). The median FLZ susceptibility for isolates was >32 μg/mL (Fig. 4C), which was higher than for the Sp1 population (2 μg/mL; SI Appendix, Fig. S5A). Sequencing of MRR1 from nine isolates revealed that the fixed SNPs present in all Sp1 and BAL isolates relative to other genomes, such as ATCC 42720, were still present in Sp2 isolates (SI Appendix, Fig. S4), indicating a single-strain background was present at all three time points over these 10 mo. MRR1 alleles in Sp2 isolates encoded Mrr1 variants different from those found in the Sp1 and BAL populations (Mrr1-K922E, K922E+N459H, F1123V, F1123Y, and E1122D+F1123L; Fig. 4C), suggesting that heterogeneity in the population either arose a second time or that multiple MRR1 alleles persisted despite FLZ treatment. We propose that while factors other than FLZ exposure lead to heterogeneity in MRR1 and thus a range of FLZ susceptibilities, the presence of isolates with high Mrr1 activity enabled the persistence of C. lusitaniae and perhaps the emergence of an even more resistant population upon FLZ treatment. It was interesting to observe three different variants of F1123 and additional alleles with two nonsynonymous changes within MRR1.
Because routine clinical microbiological assessment of drug resistance profiles involves the analysis of only one or a few representative isolates from a clinical sample, it was not surprising that the small percentage (10–20%) of FLZR isolates within the BAL and Sp1 populations escaped detection. To aid in the identification of drug-resistant subpopulations, we propose the use of pools of isolates in MIC assays. The FLZ MIC for U04 (16–32 μg/mL) in a 24-h-endpoint assay was similar to that obtained for a 9:1 mixture of FLZS (U05, MRR1L1191H+Q1197*) and FLZR (U04, MRR1Y813C) isolates, a U04 culture with a starting inoculum comparable to that in the 9:1 mixture (U04 10%), and a complex mixture of all UL isolates (Fig. 4D). Although it is surprising that a small subpopulation of drug-resistant isolates can be assessed by MIC within 24 h, further analysis of growth kinetics in the presence of FLZ showed that small resistant subpopulations within mixed populations, as in the 9(U05):1(U04) and mixture of all UL isolates samples, became detectable within 18 h (SI Appendix, Fig. S10A). Additionally, after 18 h of growth in FLZ, the culture starting at 9(U05):1(U04) was predominantly composed of U04 (MRR1Y813C) cells (SI Appendix, Fig. S10 B and C). Thus, pooled analysis of isolates may be a method by which we can detect the presence of drug-resistant subpopulations before or early on in treatment, thereby decreasing the incidence of treatment failure.
Discussion
Understanding fungal population structure within chronic infections, and awareness of the potential for the development of antifungal resistance in the absence of selection by drug, may improve individual treatment strategies. Furthermore, the common practice of analyzing only one or two isolates per infection (37–39) or per timepoint within a patient (12, 40) may not be sufficient to appreciate the phenotypes and genotypes within a large population. The analysis of drug resistance at a population level may also be important for other fungal pathogens as azole-resistance is found to be heterogeneous among isolates of Aspergillus fumigatus and C. albicans both in subjects without recent triazole treatment (41) as well as in individuals undergoing active azole treatment (42).
Azole antifungals are widely used for treatment and prophylaxis (43–45) and act by inhibiting Erg11, an enzyme essential for ergosterol biosynthesis (46). In prior reports of FLZR C. albicans, Candida parapsilosis and C. lusitaniae, the strains were isolated from patients who had received FLZ therapy (12, 40, 47). In cases where increased FLZ resistance is due to activation of Mrr1, only one mutated allele was found per patient, in contrast to the 17 alleles found here. The stable genomic diversification observed here, resulting in the up-regulation of MDR1 through hyperactivation of Mrr1, was the main driver of heterogeneous levels of FLZ resistance between isolates. Although we identified one chromosomal duplication that included ERG11, other mechanisms of FLZ resistance including increased activity of the Tac1 transcription factor to up-regulate expression of CDR1/2-encoded ABC transporters and mutations that alter FLZ binding to Erg11 were not observed (reviewed in ref. 48).
Determining what stimuli within chronic infections can contribute to the selection for antifungal resistance, either through MRR1 or other factors, will enable strategies to decrease resistance development. For example, coinfection with phenazine-producing P. aeruginosa or chronic oral colonization (where Hst 5 is abundant) may be risk factors for emergent FLZR isolates. Fungal species frequently cocolonize with bacteria and thus the effects on drug resistance may be widespread. Here, we propose that the diversity of the C. lusitaniae population may have been a consequence of the long duration of the infection, spatial complexity within the lung, and the presence of diverse selective pressures imposed by the immune system or coinfecting microbes. These factors could have contributed to both the repeated selection for and persistence of cells with different Mrr1 variants, which led to heterogeneous drug resistance within a single infection. Other chronic infections, such as those associated with bronchiectasis, oropharyngeal candidiasis, biofilms on indwelling artificial surfaces, and fungus balls may also have sufficient temporal and spatial complexity to result in heterogeneous populations. Chronic colonization of host environments that promote the selection for isolates with azole resistance have the potential to contribute to the initial evolution of epidemic multidrug-resistant strains, such as those that arose in C. auris (14).
In light of the potential benefits of high Mrr1 activity in the CF lung, the presence of Mrr1 variants with C-terminal truncations and low Mrr1 activity in this in vivo population is perplexing. These findings may indicate that there are differences between Mrr1 activity assessment in vitro and activity in vivo, or that there is a cost for Mrr1 activity in some genetic backgrounds. Although we focused on the heterogeneity in MRR1 and its link to the heterogeneous levels of FLZ resistance within the infecting population, these isolates and data may provide insight into the activities of different naturally occurring Mrr1 variants as well as other genetic changes that occur in this naturally evolved yeast population. For example, the genome data from the single and pooled isolates from the Sp1 and the UL and LL BAL samples revealed SNPs in genes encoding proteins with similarities to bile acid carriers, proteins involved in calcium homeostasis, and proteins with domains associated with signaling or protein stability. Future studies on serial samples from chronic infections may reveal other loci that enable opportunistic pathogens to adapt to the human host or influence drug treatment.
Materials and Methods
Methods describing the growth conditions, genome sequencing and analysis, transcript sequencing and analysis, mutant construction, mating experiments, drug susceptibility assays, in vitro evolution, Hst 5 sensitivity assay, zone of inhibition by P. aeruginosa, and statistical analysis are described in detail in SI Appendix, Supplemental Materials and Methods. Data and code availability statements are also included in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sven Willger (Geisel School of Medicine at Dartmouth) for helpful discussions regarding RNA-Seq data analysis and Richard Bennett for providing strains. This work was supported by NIH Grant R01 AI127548 (to D.A.H. and J.E.S.) from the National Institute of Allergy and Infectious Disease and National Institute of General Medical Sciences (NIGMS) of the NIH Awards T32GM008704 and AI133956 (to E.G.D.). Support for the BALs was provided by Grant R01 HL122372-01 (to A.A.). This work was also supported by a pilot grant (to A.A. and D.A.H.) from the Cystic Fibrosis Foundation Research Development Program (STANTO16G0 and STANTO19R0). The Cystic Fibrosis Research Development Program and NIGMS of the NIH as P20GM103413 supported the Translational Research Core at Dartmouth. Sequencing services and specialized equipment was provided by the Molecular Biology Shared Resource Core at Dartmouth, NCI Cancer Center Support Grant 5P30CA023108-37. Data analyses were performed on the Univeristy of California, Riverside High-Performance Computational Cluster supported by NSF Grant DBI-1429826 and NIH Grant S10-OD016290. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Reads for genome sequences and RNA sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. SRP133092). Data are also available through GitHub (available at https://github.com/stajichlab/C_lusitaniae_popseq). All sequence files related to this work can be found in NCBI (BioProject PRJNA433226). Whole Genome Shotgun project de novo assemblies are deposited in NCBI (accession nos. QOBD00000000–QOBX00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807698115/-/DCSupplemental.
References
- 1.Bell BG, Schellevis F, Stobberingh E, Goossens H, Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect Dis. 2014;14:13. doi: 10.1186/1471-2334-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Rie A, et al. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desnos-Ollivier M, et al. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. MBio. 2010;1:e00091-10. doi: 10.1128/mBio.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao CY, et al. Heteroresistance of Helicobacter pylori from the same patient prior to antibiotic treatment. Infect Genet Evol. 2014;23:196–202. doi: 10.1016/j.meegid.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Sun G, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherrard LJ, et al. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One. 2017;12:e0172179. doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin X, et al. Heterogeneous antimicrobial susceptibility characteristics in Pseudomonas aeruginosa isolates from cystic fibrosis patients. MSphere. 2018;3:e00615-17. doi: 10.1128/mSphere.00615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung H, et al. Global and local selection acting on the pathogen Stenotrophomonas maltophilia in the human lung. Nat Commun. 2017;8:14078. doi: 10.1038/ncomms14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins JL, Baddour LM. Candida lusitaniae infections in the era of fluconazole availability. Clin Infect Dis. 2003;36:e14–e18. doi: 10.1086/344651. [DOI] [PubMed] [Google Scholar]
- 10.Favel A, et al. Colony morphology switching of Candida lusitaniae and acquisition of multidrug resistance during treatment of a renal infection in a newborn: Case report and review of the literature. Diagn Microbiol Infect Dis. 2003;47:331–339. doi: 10.1016/s0732-8893(03)00094-4. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson BJ, Lewis RE, Kontoyiannis DP. Candida lusitaniae fungemia in cancer patients: Risk factors for amphotericin B failure and outcome. Med Mycol. 2008;46:541–546. doi: 10.1080/13693780801968571. [DOI] [PubMed] [Google Scholar]
- 12.Asner SA, Giulieri S, Diezi M, Marchetti O, Sanglard D. Acquired multidrug antifungal resistance in Candida lusitaniae during therapy. Antimicrob Agents Chemother. 2015;59:7715–7722. doi: 10.1128/AAC.02204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen XX, et al. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 2016;6:3927–3939. doi: 10.1534/g3.116.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart SR, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan DA, et al. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One. 2016;11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziesing S, Suerbaum S, Sedlacek L. Fungal epidemiology and diversity in cystic fibrosis patients over a 5-year period in a national reference center. Med Mycol. 2016;54:781–786. doi: 10.1093/mmy/myw035. [DOI] [PubMed] [Google Scholar]
- 17.Odds FC, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orenga S, James AL, Manafi M, Perry JD, Pincus DH. Enzymatic substrates in microbiology. J Microbiol Methods. 2009;79:139–155. doi: 10.1016/j.mimet.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neafsey DE, et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010;20:938–946. doi: 10.1101/gr.103911.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Ribot JL, et al. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunkel N, Blass J, Rogers PD, Morschhäuser J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol. 2008;69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branco J, et al. Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob Agents Chemother. 2015;59:6629–6633. doi: 10.1128/AAC.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favel A, et al. Susceptibility of clinical isolates of Candida lusitaniae to five systemic antifungal agents. J Antimicrob Chemother. 2004;53:526–529. doi: 10.1093/jac/dkh106. [DOI] [PubMed] [Google Scholar]
- 25.MacPherson S, et al. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother. 2005;49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell. 2004;3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 28.Reboutier D, et al. Combination of different molecular mechanisms leading to fluconazole resistance in a Candida lusitaniae clinical isolate. Diagn Microbiol Infect Dis. 2009;63:188–193. doi: 10.1016/j.diagmicrobio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Hiller D, Sanglard D, Morschhäuser J. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother. 2006;50:1365–1371. doi: 10.1128/AAC.50.4.1365-1371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morschhäuser J, et al. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirsching S, Moran GP, Sullivan DJ, Coleman DC, Morschhäuser J. MDR1-mediated drug resistance in Candida dubliniensis. Antimicrob Agents Chemother. 2001;45:3416–3421. doi: 10.1128/AAC.45.12.3416-3421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puri S, Edgerton M. How does it kill?: Understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell. 2014;13:958–964. doi: 10.1128/EC.00095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampe IAI, Friedman J, Edgerton M, Morschhäuser J. An acquired mechanism of antifungal drug resistance simultaneously enables Candida albicans to escape from intrinsic host defenses. PLoS Pathog. 2017;13:e1006655. doi: 10.1371/journal.ppat.1006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price KE, et al. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome. 2013;1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr JR, et al. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson R, et al. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard SJ, et al. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009;15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nucci M, et al. Latin American Invasive Mycosis Network Epidemiology of candidemia in Latin America: A laboratory-based survey. PLoS One. 2013;8:e59373. doi: 10.1371/journal.pone.0059373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tadec L, et al. Epidemiology, risk factor, species distribution, antifungal resistance and outcome of candidemia at a single French hospital: A 7-year study. Mycoses. 2016;59:296–303. doi: 10.1111/myc.12470. [DOI] [PubMed] [Google Scholar]
- 40.White TC, Pfaller MA, Rinaldi MG, Smith J, Redding SW. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 1997;3:S102–S109. doi: 10.1111/j.1601-0825.1997.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim SH, et al. Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor Nrg1 as a mechanism of pathogen adaptation. PLoS Pathog. 2015;11:e1005308. doi: 10.1371/journal.ppat.1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez M, et al. Heterogeneous mechanisms of azole resistance in Candida albicans clinical isolates from an HIV-infected patient on continuous fluconazole therapy for oropharyngeal candidosis. J Antimicrob Chemother. 2002;49:515–524. doi: 10.1093/jac/49.3.515. [DOI] [PubMed] [Google Scholar]
- 43.Ceesay MM, et al. Triazole antifungals used for prophylaxis and treatment of invasive fungal disease in adult haematology patients: Trough serum concentrations in relation to outcome. Med Mycol. 2016;54:691–698. doi: 10.1093/mmy/myw031. [DOI] [PubMed] [Google Scholar]
- 44.Lass-Flörl C. Triazole antifungal agents in invasive fungal infections: A comparative review. Drugs. 2011;71:2405–2419. doi: 10.2165/11596540-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 45.Pappas PG, et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2016;62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med. 2014;5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, et al. Development of fluconazole resistance in a series of Candida parapsilosis isolates from a persistent candidemia patient with prolonged antifungal therapy. BMC Infect Dis. 2015;15:340. doi: 10.1186/s12879-015-1086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol. 2017;71:753–775. doi: 10.1146/annurev-micro-030117-020345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.