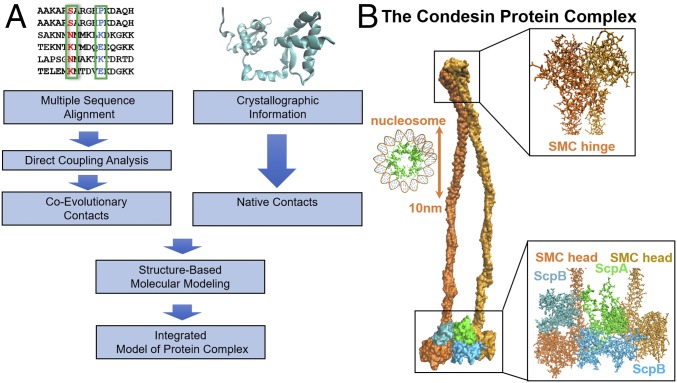
Fig. 1.
Schematic of our integrative computational approach together with the resulting proposed structure for the prokaryotic condensin complex. (A) We obtain MSAs for the protein families of SMC, ScpA, and ScpB from Pfam (41). Using these sequence data as input, DCA is used to infer intraprotein and interprotein coevolving contacts. Known crystal structures of the condensin subunits are obtained from the PDB (27, 28). To obtain the complete structure of the condensin complex, we use both coevolutionary and crystallographic residue contacts as constraints in MD simulations. (B) The proposed structure for whole condensin complex, composed of 2,924 residues at atomic resolution. A single-ring structure is consistent with all of the available information, both structural and coevolutionary. A nucleosome (PDB ID code 5T5K) is shown on the right-hand side to illustrate the scale of the figure.