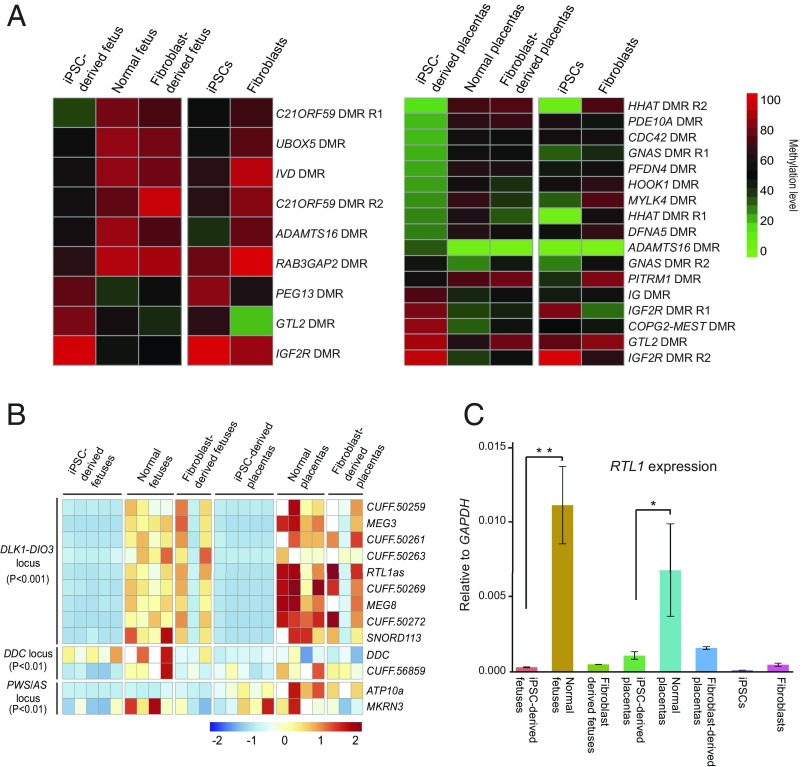
Fig. 5.
Aberrant methylation and gene expression of imprinted regions in iPSC-derived fetus and placenta. (A) Heat map of DMRs in iPSC-derived fetus/placenta compared with normal fetus/placenta in imprinted loci. The imprinted loci include the candidate imprinted regions and homologous imprinted regions inferred from mouse. (B) Heat map of iDEGs (known plus candidate) in different tissues. The list of iDEGs includes iDEGs between iPSC-derived fetus and normal fetus, and iDEGs between iPSC-derived placenta and normal placenta. DEGseq adjusted P value is used for detecting DEGs. (C) RTL1 expression measured by strand-specific quantitative PCR. The iPSC-derived fetus/placenta show significant lower RTL1 expression than the normal fetus/placenta (n = 3, *P < 0.05; **P < 0.01, two-tailed Student’s t test).