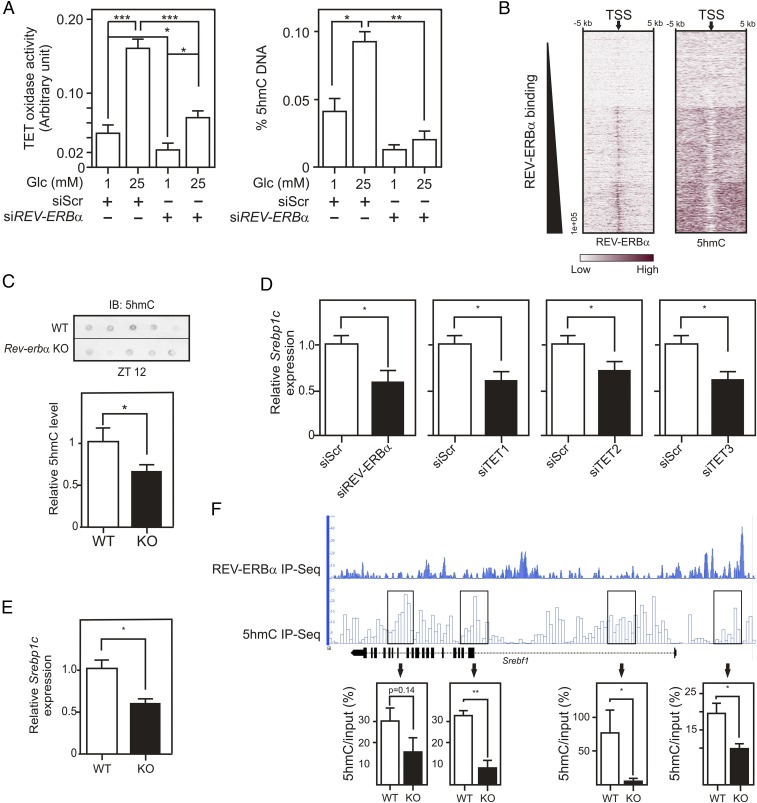
Fig. 6.
Nuclear REV-ERBα controls nuclear OGT and TET activities and affects DNA hydroxymethylation. (A) TET oxidase activities (Left) and 5hmC levels (Right) in HepG2 nuclear protein extracts or in genomic DNA, respectively. Cells were transfected with siRNA (control or REV-ERBα) and incubated at 1 or 25 mM Glc. (B) Heatmaps of REV-ERBα and 5hmC signal intensities in clusterized Gencode TSS. Gencode TSS (arrow) were aligned and extended 5 kb on each side. (C) 5hmC levels in Rev-erbα+/+ (WT) and Rev-erbα−/− (KO; n = 5–6) mouse livers. (D) Relative SREBP1C gene expression determined by RT-qPCR on siRNA (Scr, REV-ERBα, TET1, TET2, or TET3)-transfected HepG2 cells. (E) Relative Srebp1c gene expression in liver from Rev-erbα+/+ or Rev-erbα−/− mice (ZT12). (F) Representation of REV-ERBα chromatin occupancy and hydroxymethylated region localization at the mouse hepatic Srebf1 locus (Upper). Srebf1 hydroxymethylation was quantified by hMeDIP-qPCR (Bottom) in hepatic genomic DNA from Rev-erbα+/+ (n = 2) or Rev-erbα−/− mice (n = 2). Histogram represents mean ± SEM. The statistical significance of differences was assessed by a two-way ANOVA followed by a Bonferroni post hoc test (A) or by a t test. *P < 0.05, **P < 0.01, ***P < 0.001.