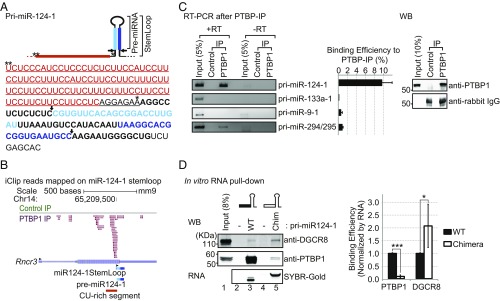
Fig. 2.
PTBP1 binds to a CU-rich segment in pri-miR-124-1. (A) The sequence of pri-miR-124-1 with its secondary structure and sites of DROSHA cleavage (arrows) are diagrammed (Top). The miR-124 duplex strands resulting from DROSHA and DICER cleavage are indicated in light and dark blue, with the final miR-124 in dark blue. Asterisks mark nucleotides 1 and 107 upstream from miR-124 stem–loop, respectively. The CU-rich segment extending from 107 nt to 8 nt upstream of the stem–loop is in red. Sequences switched with pri-miR-1a-2 to form the chimeric RNAs in D are underlined. (B) Genome browser view of PTBP1 iCLIP tags (PTBP-IP, magenta) and FLAG-rabbit (Control-IP, green) from mESCs aligned to the miR-124-1 host gene (Rncr3, AK044422). The stem–loop of miR-124-1, pre-miR-124-1, and the CU-rich segment are indicated (Bottom). Black arrowheads indicate 5′ to 3′ direction of each of RNA. (C) Immunoprecipitation of pri-miR-124-1 with PTBP1. PTBP1 was immunoprecipitated from mESC (PTBP1-IP) and bound RNA was extracted and assayed for pri-miRNAs by RT-PCR. Rabbit IgG served as a negative control (Control-IP). PCR in the absence of reverse transcription (−RT) served to control for genomic DNA contamination. Representative gel images from three biological replicates are shown (Left). RNA bound to PTBP1-IP was compared with 5% of the input RNA (Middle); error bars are SEM. Immunoprecipitated targets were checked by Western blot (Right). (D) The CU-rich segment is required for the interaction between PTBP1 and pri-miR-124-1. A chimeric miRNA had the CU-rich segment of pri-miR-124-1 (black box) replaced with the equivalent sequence from pri-miR-1a-2 (open box). WT and chimeric (Chim) pri-miR-124-1 were transcribed in vitro and hybridized to biotinylated adaptors immobilized on streptavidin beads. Beads carrying biotinylated adapters alone served as negative controls (lanes 2 and 4). Pri-miRNA–bound beads were incubated with mESC total cell extract in 2 mM EDTA to inhibit DROSHA cleavage. After washing, bound PTBP1 and DGCR8 were detected by Western blot and the bound RNA with SYBR-Gold (Left). Representative gel images are from three biological replicates. Protein bound to each RNA was quantified, normalized to the amount of bait RNA, and further normalized to WT RNA in each replicate (Right) (n = 3 biological replicates; Student’s t test; *P ≤ 0.05, ***P ≤ 0.001; error bars are SEM).