Figure 3.
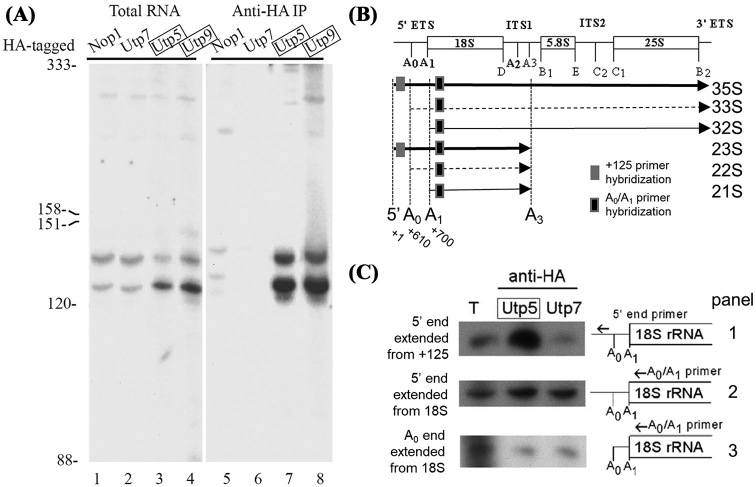
The short 5΄ ETS rRNA transcripts are associated with the t-Utps. (A) The indicated proteins were HA-tagged by chromosomal integration. Nop1 is a core box C/D snoRNA protein and Utp7 is not a t-Utp; Utp5 and Utp9 are t-Utps (boxed). RNAs co-immunoprecipitated by HA-tagged proteins were probed with an oligonucleotide complementary to nt 1–24 of the pre-rRNA (3΄ start oligo; Table 1). The blot was re-probed for tRNA-Tyr, 5S rRNA, 5.8S rRNA and the U3 snoRNA, and their sizes are indicated. Total RNA represents 10% of input from each immunoprecipitation (lanes 1–4). RNAs associating with Nop1-HA, Utp7-HA, Utp5-HA and Utp9-HA were analyzed in lanes 5–8. (B) Different 18S pre-rRNA species generated from different endonucleolytic events. U3 snoRNA dependent cleavages are in bold and the nucleotide is noted under. Internal transcribed spacer (ITS) 1 and 2 separate the mature rRNAs while the external transcribed spacers (ETS) flank the pre-rRNA. 5' ETS primer hybridization are noted in gray boxes while the A0/A1 primer hybridization is a black box. (C) Primer extensions of RNA immunoprecipitated from HA-tagged Utp7 and Utp5 and total RNA (T) were carried out with indicated primers.