Figure 4.
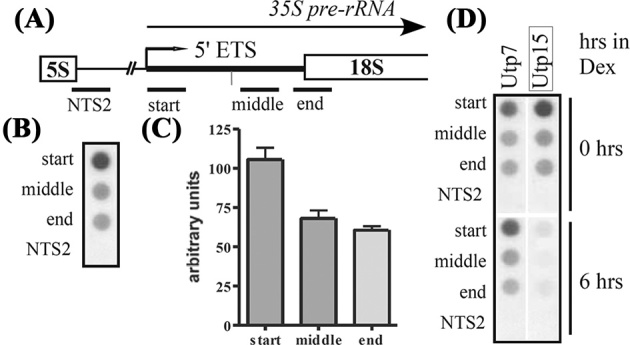
The short 5΄ ETS rRNA transcripts are actively transcribed and are dependent on t-Utps. (A) Underlines indicate probes used in the transcriptional run-on assays. The non-transcribed spacer (NTS2) probe is approximately 1 kb upstream from the pre-rRNA transcription start site. The start probe spans from 1 to 194 nucleotides of the 5΄ external transcribed spacer (ETS) pre-rRNA. The middle probe spans 350–631 nucleotides of the 5΄ETS pre-rRNA. The end probe spans 687–945 nucleotides, from within the 5΄ETS pre-rRNA and extending into the 18S rRNA. The A0 cleavage site is marked with a vertical line. (B) Radiolabeled RNA transcripts from transcriptional run-on assay were used to hybridize to membranes containing the four probes to the rDNA. (C) Quantitation of transcriptional run-on assay in Figure 3B. The signal was normalized to NTS2 and standard error is shown from three independent assays. (D) Transcriptional run-on assays were carried out on GAL: HA-UTP strains. Radiolabeled RNA transcripts from cells depleted of Utp7 and Utp15 for zero and six hours by growth in dextrose-containing media were used to probe blots containing fragments of rDNA.
