Figure 5.
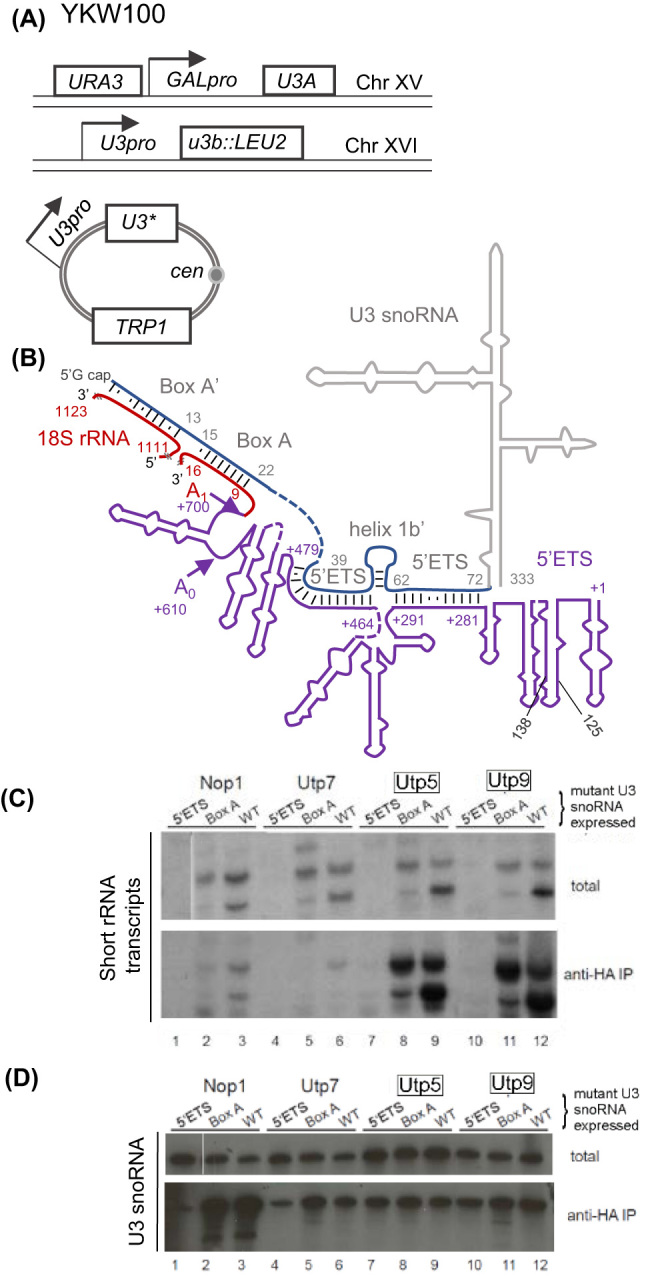
Early 5΄ETS pre-rRNA-U3 snoRNA interaction is required for the production of short transcripts and association with the other Utps with the U3 snoRNA. (A) Schematic of yeast strain YKW100 containing U3B on chromosome XVI replaced with LEU2, U3A placed under the control of the GAL promoter. Different plasmids containing U3 mutants (U3*) are expressed under the endogenous U3 promoter. (B) Schematic of the U3 snoRNA and 5’ETS of the rRNA basepairing. Sequences comprising the mature 18S rRNA are red while the 5΄ETS is in purple and relevant nucleotides are in matching colors numbering begins at the first nucleotide of that region (ETS starts at +1). Canonical basepairing is noted a black dash and other nucleotide interactions are a dot. Regions of the U3 snoRNA interacting with the pre-rRNA are blue and the body of the U3snoRNA not directly interacting with the pre-rRNA is grey. Cleavage sites are noted with arrows. (C) RNA was co-immunoprecipitated from yeast with C-terminal HA-tagged Nop1, Utp7, Utp5 and Utp9 expressing mutant U3 snoRNA (5΄ETS or Box A binding sites) or wild-type U3 snoRNA (WT). Total RNA represented 5% of lysate immunoprecipitated. Northern blots were probed with the start oligo complementary the 5΄ end of the pre-rRNA. t-Utps were boxed. (D) Northern blot in part C was reprobed to detect co-immunoprecipitated U3 snoRNA expressed from a plasmid, extracted yeast expressing C-terminal tagged Nop1, Utp7, Utp 5 and Utp9. t-Utps are boxed.
