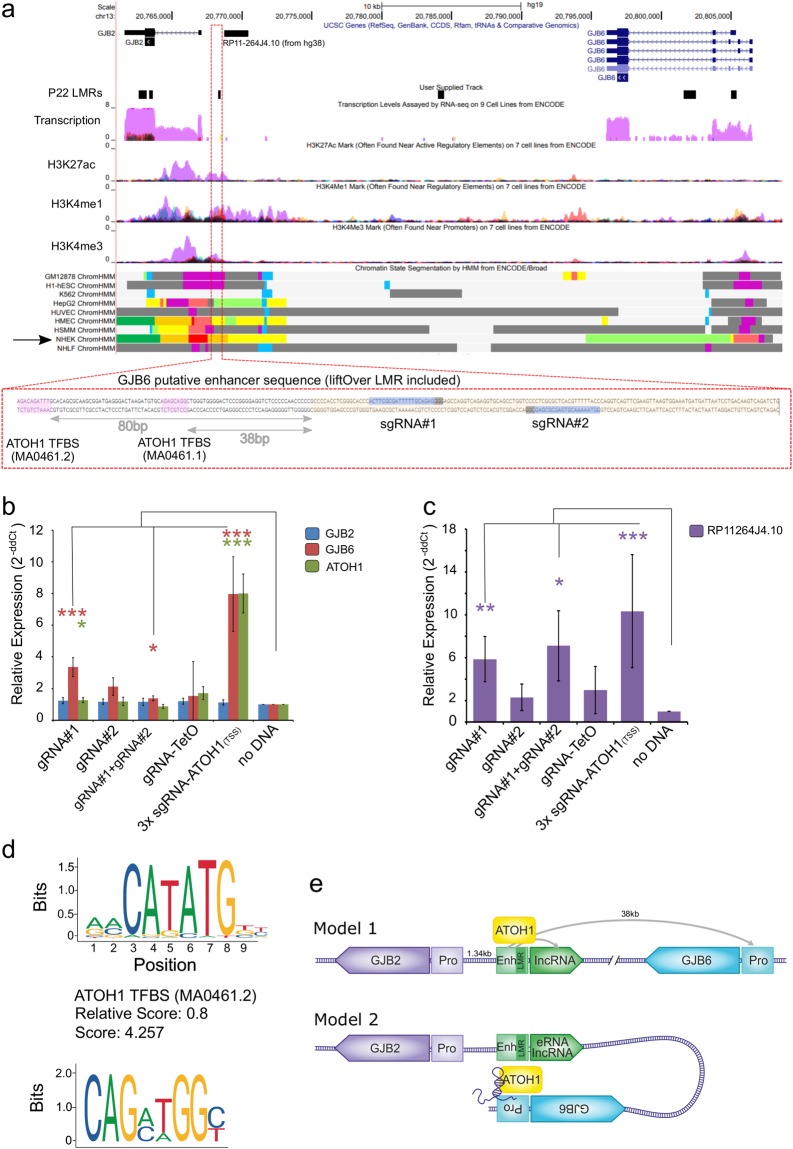
Figure 7.
CRISPR-on modulation of GJB6 and non-coding lncRNA RP11-264J4.10 expression via a putative enhancer. (a) UCSC browser snapshot presenting the candidate enhancer lifted over from mouse (mm10) to human (hg19) (track: “P22 lifted LMRs”, black box indicators); additional information derived from ENCODE regulation hub is presented to support the characterization of the candidate sequence as an enhancer (track: ‘transcription’ - purple signal = NHEK cell line, track: ‘NHEK ChromHMM’ – orange indicating ‘Active enhancer’). Below, zoomed view of the candidate sequence, with the location of the lifted-over LMR, gRNA targets and ATOH1 TFBS indicated. (b, c) Expression of GJB2, GJB6 and ATOH1 (b) and RP11-264J4.10 ncRNA (c) measured by qRT-PCR (normalized to GAPDH, compared to ‘no DNA’, n = 4–5, ***p < 0.01, *p < 0.05). (d) ATOH1 transcription factor binding motif found at the candidate enhancer, score is calculated by JASPAR. (e) Suggested mechanism of GJB6 regulation by the putative enhancer, shown in two models. Model 1 depicts a physical distal interaction between the GJB2 proximal putative enhancer (yellow box) and the GJB6 promoter (light orange box) mediated by ATOH1 and/or the lncRNA (purple). Model 2 demonstrates the folding of the chromatin, linking the putative enhancer with the GJB6 promoter, mediated by ATOH1 and/or the lncRNA.