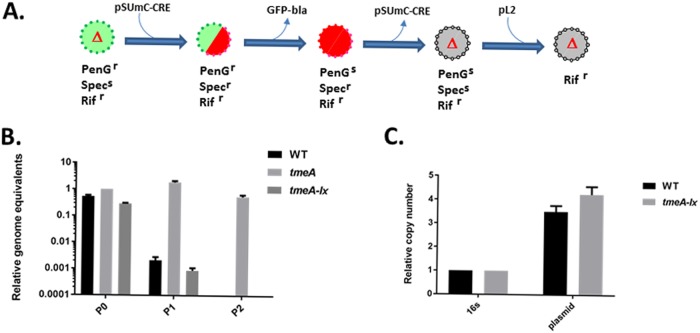
FIG 3.
Construction of a markerless Chlamydia tmeA mutant. (A) Schematic representation of the strategy used to create a markerless tmeA mutant. Each intermediate is depicted with fluorescent qualities (green, GFP+; red, mCherry+; gray, no fluorescence) and antibiotic sensitivities. (B) McCoy cell cultures were infected with equal IFUs of WT, L2 tmeA, or L2Rif tmeA-lx C. trachomatis and serially passaged every 24 h in medium containing PenG. At each passage, DNA was harvested for quantitative real-time PCR to determine genome equivalents based on chlamydial 16S rRNA. (C) qPCR-based comparison of endogenous pL2 (plasmid) copy number in WT or tmeA-lx mutant chlamydiae relative to chlamydial 16S rRNA. DNA was harvested from infected McCoy cells at 24 hpi. PenGr, penicillin G resistant; PenGs, penicillin susceptible; Rifr, rifampin resistant; Specs, spectinomycin susceptible; Specr, spectinomycin resistant.