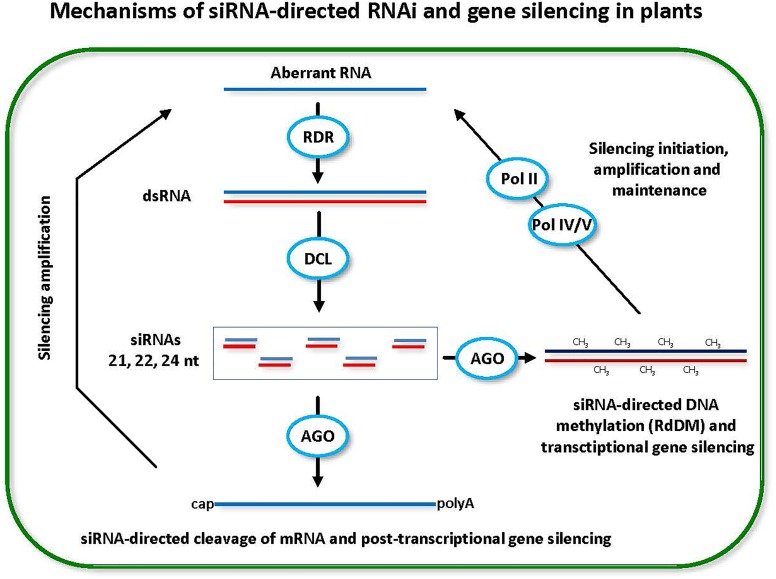
FIGURE 1.
Small interfering RNA (siRNA)-directed RNA interference (RNAi) and gene silencing in plants. Double-stranded RNA (dsRNA) is a trigger of RNAi and gene silencing at both post-transcriptional and transcriptional levels (PTGS and TGS). Dicer-like (DCL) family enzymes catalyze processing of dsRNA into siRNA duplexes. One of the duplex strands gets associated with an Argonaute (AGO) family protein and the resulting RNA-induced silencing complex targets cognate genes for PTGS through target mRNA cleavage and/or TGS through target DNA methylation (CH3). Positive feedback loops reinforce both PTGS and TGS. In PTGS, the mRNA cleavage products (aberrant RNAs) are converted by RNA-dependent RNA polymerase (RDR) activity into dsRNA precursors of siRNAs. In TGS, the methylated DNA and the target (to-be-methylated) DNA are transcribed by Pol IV and Pol V, respectively. The Pol IV transcripts are converted by RDR activity into dsRNA precursors of siRNAs, while the Pol V transcripts serve as scaffolds that interact with siRNA-AGO complexes and recruit DNA methyltransferase that mediates de novo RNA-directed DNA methylation (RdRM). Pol II is likely involved in initiation of RdDM and TGS by producing aberrant transcripts that are recognized by RDR and converted into dsRNA precursors of siRNAs.