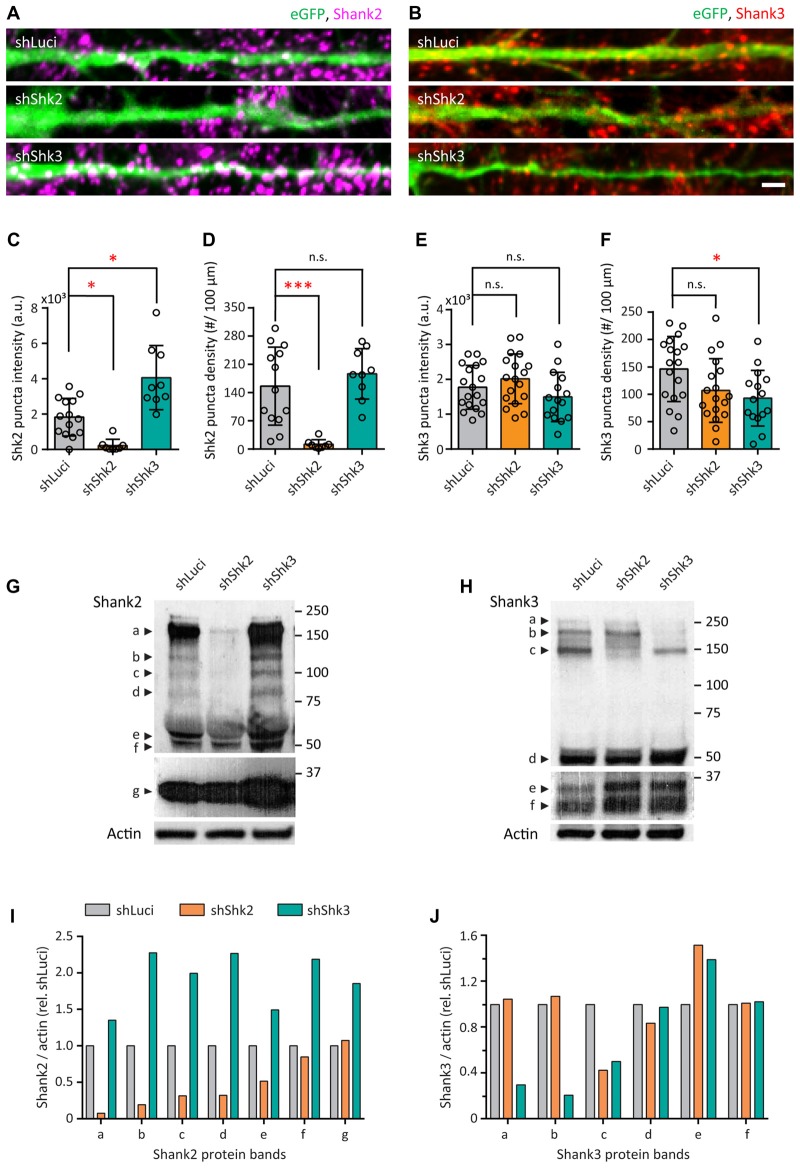
Figure 10.
Short-hairpin RNA (shRNA)-mediated knockdown of Shank2 and Shank3. (A,B) Straightened dendrites of transfected hippocampal neurons expressing a bicistronic construct with enhanced green fluorescent protein (eGFP; green) and shRNA hairpins for shLuciferase (shLuci, top), shShank2 (shShk2, middle) or shShank3 (shShk3, bottom) and immunostained for Shank2 (magenta; A) or Shank3 (red; B) after 14 DIV. Scale bar: 5 μm. (C–F) Summary graphs showing quantification of Shank2 (C,D) and Shank3 (E,F) puncta intensity and density (mean ± SEM) for different knockdown conditions (Kruskal-Wallis one-way ANOVA with Dunn’s correction post hoc multiple comparisons, Shank2 staining: shLuci, N = 14; shShk2, N = 8; shShk3, N = 9 dendrites from 8 to 12 neurons from three culture preps. Shank3 staining: shLuci, N = 18; shShk2, N = 18; shShk3, N = 15 dendrites from 8 to 12 neurons from three culture preps; n.s. p ≥ 0.05, *p < 0.05, ***p < 0.001).(G,H) Western blots of cellular lysates from dissociated hippocampal neurons infected with lentivirus for shLuci, shShk2 or shShk3 immunoblotted for Shank2 (G) or Shank3 (H) antibodies. Protein molecular weights are indicated at the right in kDa. Different isoforms are labeled at the left. Top two panels in (G,H) show different exposures of the same films for optimal visualization of different isoforms. Actin was used as a loading control (bottom panels in G,H). (I,J) Quantification of Shank2 (I) or Shank3 (J) protein levels for different isoforms in hippocampal neurons for different knockdown conditions from two cultures per condition. “Shank/actin (rel. shLuci)” refers to the expression level of Shank2 or Shank3 normalized to actin and then to shLuci control.