Abstract
Neuroblastoma is a neural crest-derived tumor that accounts for 7–10% of all malignancies in children and ~15% of all childhood cancer-associated mortalities. Approximately 50% of patients are characterized as high-risk (HR) and have an overall survival of <40% at 5 years from diagnosis. HR patients with unfavorable prognosis exhibit several structural copy number variations (CNVs), whereas localized tumors belonging to patients in the low- and intermediate-risk classes, have favorable outcomes and display several numerical CNVs. Taken together these results are indicative of chromosome instability (CIN) in neuroblastoma tumor cells. The present review discusses multiple aspects of CIN including methods of measuring CIN, CIN targeting as a therapeutic strategy in cancer and the effects of CIN in neuroblastoma development and aggressiveness with particular emphasis on the CIN gene signature associated with HR neuroblastoma patients.
Keywords: neuroblastoma, chromosome instability, chromosome instability signature, tumorigenesis
1. Introduction
Neuroblastoma (NB) is a pediatric cancer with the highest incidence during pre-scholar age. Patient can onset with localized or metastatic tumor. High-risk (HR) patients usually are older than one year of age with a metastatic tumor involving bone marrow, liver and skeletal bone (1). The overall survival (OS) of HR patients at 5-years from diagnosis is near 40%. On the contrary, patients with localized tumors, usually fall in the Low-Risk or Intermediate-Risk group, and have a 5-years OS ranging between 75 and 98% (2).
Tumor cells show several gross cytogenetic abnormalities. The most aggressive tumor in children older than 1 years of age, shows numerous copy number variations (CNVs) such as deletion, gains and gene amplification with homogenously staining region and/or double min and/or entire chromosome extra-copies (3). Two type of CNVs can occur in tumor cells: Structural CNVs that shows structural chromosome changes such as deletion, amplification and gross chromosome rearrangement, and numerical CNVs that involves gain or loss of whole chromosomes (4). Tumor aggressiveness has been observed strongly associated with such structural CNVs whereas localized tumors, belonging to patients at low- and intermediate-risk, are less aggressive and they have numerical CNVs, prevalently.
Recently, another phenomena observed in NB tumors is the chromotripsys at chromosome 5, particularly (5). Moreover, NB tumor cells show different ploidy features: near-diploidy and near-tetraploidy are observed in HR tumors characterized by structural abnormalities, instead near-triploidy cells are usually present in tumors of low-risk patient (6); these tumors show extra-chromosomes and few structural chromosome abnormalities. Consistently, patients with the near-triploidy tumor cells have favorable outcome with a good OS, whereas unfavorable prognosis is observed in patients who have near-diploidy or near-tetraploidy tumors (7,8). All together these findings show evidence of chromosome instability (CIN) in NB cells.
2. CIN and tumorigenesis
In the past years, enormous interest among the scientists has stimulated the search for gene mutations as causative of tumorigenesis (9–11). The development of next generation technique (NGS) has greatly improved the understanding of several cancer molecular mechanisms, with the prospective to improve the treatment by targeting cancer mutated proteins. However, prior to the interest for gene mutation research, there was a long period dominated by the search of numerical and structural chromosomal abnormalities associated to the genesis of several tumors (12). In 1914, Boveri first suggested the hypothesis of aneuploidy in cancer, but only later the relevant role of CIN in the majority of tumors was demonstrated by Cahill et al (13) and by Heng et al (14). To date, there is a debate if the CIN is the cause of genomic instability (GIN) developing the cancer (15,16) or if CIN process is the consequence of abnormal function of mutated gene encoding for important mitotic proteins leading to tumor development (17,18).
CIN with chromosome aberrations in copy number is a form of GIN that make prone the cancer cells to acquire mutations conferring them rapid tumor progression, aggressiveness and drug-resistant phenotypes. GIN is characterized by an increased frequency of genetic alterations deregulating specific biological signaling associated with cellular cell cycle homeostasis and induces genomic diversity in cancer cells (19). This genomic chaos gives to cancer cell populations the properties to adapt themselves at the stimuli of the tumor microenvironment (20).
For long time, the chromosome missegregation together with others mechanisms such as gene mutations, chromosomal rearrangements and epigenetic factors were considered responsible for tumor growth and tumor heterogeneity. Indeed, cell heterogeneity is a hallmark of tumors lending the ability of adapting to external pressures (21,22). Furthermore, many studies are reporting the link among karyotype alterations, CIN (23) and cancer (13,24–26) strongly indicating that CIN takes part in the origin of cancer (18,27). Moreover, the role of CIN in tumorigenesis is also supported by experimental observations of persistent chromosome missegregation in tumor cell lines (28). Altogether these observations indicate the link between CIN and aneuploidy (29).
There are evidences that CIN and aneuploidy could promote tumor initiation, acquisition of drug resistance, metastasis and relapse (30) through the variation of the copy number of oncogenes/tumor suppressor genes allowing cells to adapt to environmental stimuli changes such as nutrient variation and/or hypoxia (20). Aneuploidy may induce genome instability and lead to acquisition of genetic cell heterogeneity triggering selective pressures in clones selection. Therefore, the survival of cells with CIN to the cancer treatment could be due to higher adaptive potential of these cells in which specific chromosomal aberrations confer cellular fitness advantages (31).
3. The aneuploidy paradox
Despite aneuploidy commonly occurs in many cancers where it is an indicator of tumor growth, it often leads to a reduction in the cell proliferation rate (32,33). This apparent contradiction is known as the: aneuploidy paradox. The rate of CIN determines the effect of aneuploidy on tumors; whereas low rates of CIN (missegregation of a small number of chromosomes per division) are weakly tumor promoting, higher rates of CIN (missegregation of more than five chromosomes) cause cell death and tumor suppression (34). Thus, cell death induced by CIN sufficiently high arises from an increase in the number of chromosomes missegregated per cell division. Coherently with this, Komarova described a mathematical model showing that a low rate of CIN optimizes the tumor heterogeneity and survival and that increase of CIN rate is associated with decreases tumor fitness (35). These observations suggest that increasing the rate of CIN over a critical threshold could be efficient to stop the tumor cell proliferation.
This paradoxical situation is attracting considerable attention for therapeutic purpose, although the therapeutic targeting of CIN in cancer is still at preclinical stages. Several oncogenes and tumor suppressor genes are known to localize within centrosomes, which altered function triggers centrosome abnormalities (36). Thus, there are many promising inhibitors against associated centrosome proteins and many of these drugs/compounds are being tested in preclinical models and in clinical trials. Two of the most studied centrosomal kinases with oncogenic properties are: AURKA and AURKB, and more than 30 AURK inhibitors have been developed and used in clinical studies [reviewed by (37)].
Tubulin is another important target, as this protein acts during cellular growth, division, and migration. Taxanes (paclitaxel and docetaxel) and vinca alkaloids (vinblastine, vincristine, and vinorelbine), well-known FDA-approved compounds clinically used for targeting tubulin, have been demonstrated to be successful to induce mitotic arrest (38).
Additional cancer therapy strategy that has attracting attention in recent years is synthetic lethality (39), defined as a condition in which perturbation in two different genes together results in cell death but mutation of either alone is compatible with cell life. The first clinically approved drugs designed to exploit synthetic lethality are poly(ADP-ribose) polymerase inhibitors (PARPis) (40). PARP is a nuclear protein important for recognizing DNA damage and repairing DNA single-strand breaks (SSBs). It is proposed that inhibition of PARP results in the accumulation of unrepaired SSBs that are converted into double-strand breaks during DNA replication resulting in gross GIN and cell death.
4. Methods of measuring CIN
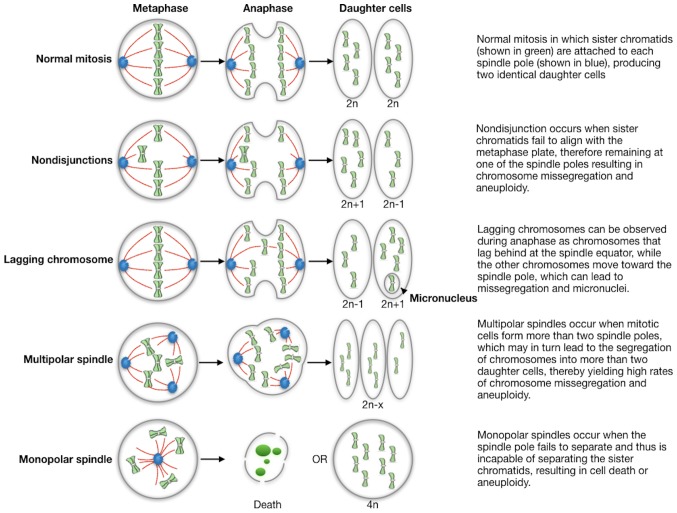
Although the evaluation of CIN rate in tumor samples is not routinely performed in the clinical setting, direct and indirect methods to measure CIN have been adopted; these methods are based on both the determination of cell-to-cell variability in chromosome number and structure within the tumor cell population, as well as on the assessment of the rate at which these chromosomal changes occur (41). Therefore, the methods are able to capture the dynamic nature of CIN. The CIN rate is directly related to the estimation of the mitotic error frequency in fixed cells or in formalin-fixed and paraffin-embedded (FFPE) tumor tissues. Some of main defects are summarized in Fig. 1: abnormalities in chromosome structure and function resulting in chromosomes that lag in anaphase or exhibit incomplete separation of sister chromatids; spindle abnormalities such as multipolar spindles and defects in cytokinesis are additional sources of abnormal chromosome segregation; finally, errors in cell cycle regulation, including delays during division and defects in cell cycle checkpoints, also leading to missegregation. However, these technical approaches are difficult to apply in tumors with a low proliferation index and in tumors in which the anaphases are not clearly observed.
Figure 1.
Schematic presenting the causes of chromosome missegregation in mitosis.
CIN is not only a consequences of compromised mitotic apparatus but it may also arise after DNA damage or as a consequence of impaired replication fork progression (42). Defective DNA damage response and repair results in chromosomal aberration such as deletions, amplifications, inversions and translocations (43).
The main method for the assessment of both numerical and structural CIN in tumor cells is the fluorescence in situ hybridization (FISH). It allows to quantify the variations in chromosome copy number across the cell population by using fluorescently labeled DNA probes that bind to centromeres of specific chromosomes. Thus, FISH evaluates the chromosomal state of hundreds of cells, inferring the rate of change in chromosome number from the cell-to-cell variability (44). Another important method is the single-cell comparative genomic hybridization (CGH) (45). This assay allows the selection of individual cancer cells based on their deviation from normal cells. However, single-cell CGH is not amenable to high-throughput analysis and it is characterized by a considerable economic burden. Moreover, flow cytometry and DNA image cytometry can be used to measure cellular DNA content through the use of dyes that bind the DNA, allowing the assessment of DNA cell cycle distribution and cellular ploidy. Then, CIN status can be measured by the stemline scatter index (SSI), which is a measure of the clonal heterogeneity of the constituent tumor cells (46). Finally, a more detailed picture of the genomic landscape can be obtained through Next Generation Sequencing (NGS) systems, which assume massively parallel sequencing techniques (47).
Recently, different models have been used to measure CIN in vivo like mouse models that were engineered to mimic genetic alteration driving CIN, mouse embryonic fibroblast (MEFs) from mouse models of genetically induced CIN (48) and organoid cultures, that allow to monitoring chromosome segregations using three-dimensional live-cell imaging (49). Measuring CIN in vivo would more accurately show the effect of CIN for instance during development, the possible role of the immune system and inter-tissue interactions. However, drawbacks of in vivo CIN measurement are the limited time available for imaging, the high cost and the relatively low rate of cell division in vivo (50).
Despite the studies performed so far, none of the methods used to study CIN is entirely satisfactory, thus novel approaches for an accurate detection and assessment of CIN will be critical both in clinical setting and to therapeutic targeting of CIN in the future.
5. CIN takes part in NB development and aggressiveness
NB is an embryonic tumor that can be present in fetus. Indeed, some newborn patients exhibit tumors after only a few days of life; usually, these patients have a very good OS. Cytogenetic analysis of tumor cells reveals a triploid DNA content with several numerical CNVs. This, of course, is in contrast with the presence of structural CNVs in tumor of HR patients older than one year of age, suggesting an evolution of tumor aggressiveness associated with CIN. The lapse time between fetus life and infant at one age of year suggest a time-dependent increase of chromosome damages (51). As mentioned above, cell replication errors and abnormal chromosome segregation during mitosis could originate the abnormal chromosome pattern triggering the cell to increase their aggressiveness. Masecchia et al (52) used a learning machine algorithm to show that numerical chromosome aberrations occurs early than structural ones.
It is to note that NB is originating from neural crest cells, a group of cells located on the neural tube and undergoing to epithelial to mesenchymal transition (EMT) during the embryonic life. These cells are migrating in the early phases of embryonic development and some of them take part to the formation of gastric ganglia and adrenal gland, two sites in which NB growths and develops. Mouse and zebrafish models have been developed demonstrating that MYCN oncogene is one of the major actors in the NB development (53). Furthermore, it has been shown that ALK and LIN28 genes can participate together MYCN oncogene to the NB tumorigenesis (54). The role of CIN in the embryonic phases of NB origin is still to clarify. It is questionable if some CIN-related genes are involved in the early phases of NB tumorigenesis. This study needs a more accurate animal model.
6. CIN gene signature and NB
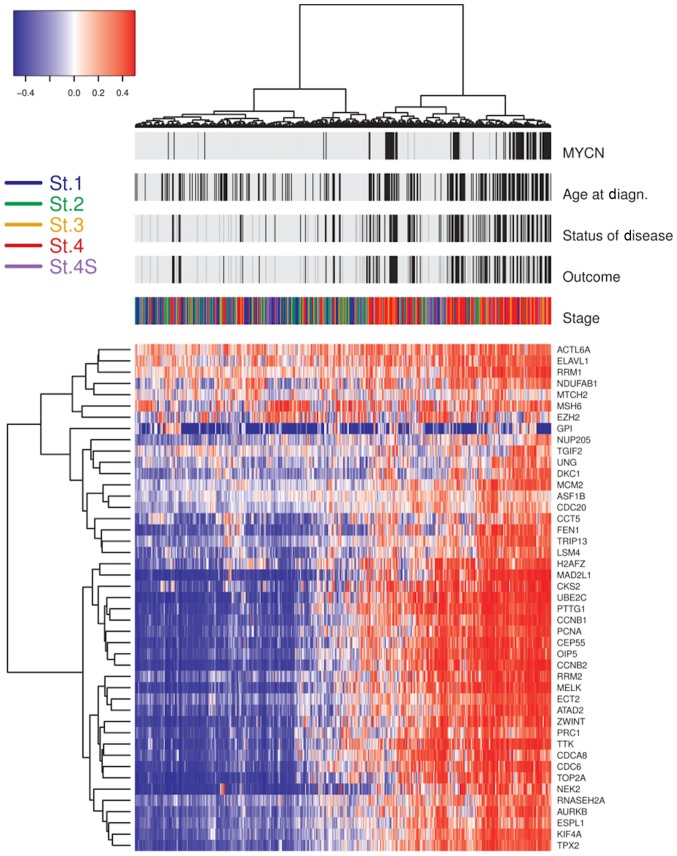
Considering the strong relationship between CIN and aneuploidy, about a decade ago, a computational method was developed to represent aneuploidy in relation to the expression of genes localized in aberrant chromosomal region (functional aneuploidy profile). Thus, the functional aneuploidy as a measure of the total status of chromosomal imbalance, was inferred using gene expression data of a given tumor (55). Carter et al (55) showed 70 genes whose expression was correlated with total functional aneuploidy in several cancer types: the CIN70 gene signature able to measure the state of karyotype and to predict clinical outcome in several human cancers. The CIN70 signature was obtained including most of genes involved in cellular processes critical for genome integrity maintenance such as DNA replication, chromosomal condensation, segregation, de-condensation and structure and genes of cell cycle, spindle apparatus and mitosis. Carter's study provided a means to assess the potential role of CIN in tumors initiation. However, in this study NB tumor was not investigated and CIN signature for NB has not been identified until now. Since this information is lacking, we explored gene expression profiles of 504 NB derived from public dataset E-MTAB-161 (EMBL ArrayExpress database). This dataset provided expression data of 45 genes out of 70 genes included in the CIN70 signature. Patients clinical information was used to define two risk groups: HR group including samples with stages 2, 3, 4 and 4s MYCN-amplified, stage 4 MYCN not amplified >12 months at diagnosis; low/intermediate-risk (LIR) group including samples with stages 2, 3 and 4S without MYCN gene amplified, stage 4 without MYCN amplification <12 months at diagnosis. We observed 31/45 genes differently expressed between HR and LIR groups (Fig. 2). These genes show association with cell-cycle regulation (CCNB1, PRC1 and TPX2) and chromosomal segregation (TPX2). Furthermore, we found overexpression of key regulators genes involving in the correct processes of chromosomes replication, regulation of chromatin status, cytokinesis and segregation; namely: AURKB, CCNB1, CCNB2, NEK2 and ZWINT and likely associated with CIN in HR-NB patients. Interestingly, we observed genes with the highest CIN score reported in Carter's study, among genes with high expression in HR-NBs, such as TPX2 and PRC1.
Figure 2.
Unsupervised clustering of 45 genes with CIN70 in NB. The heatmap of the unsupervised clustering presents 30 genes with differential expression between NB-high risk patients (stages 2, 3, 4, and 4S with MYCN gene amplification; stage 4 patients with an age at diagnosis >12 months) and low/intermediate-risk patients (stage 1, 2, 3, 4S without MYCN gene amplification). Above the heatmap are the indicated patient clinical features and the patients' stage. Black bars represent unfavorable scores and light gray bars represent favorable scores for ‘Status of Disease’ and ‘Outcome’. Black bars for the ‘Age at Diagn.’ identify cases with ages over 60 months at diagnosis. Black bars for ‘MYCN’ identify cases with amplification of MYCN gene. On the left of the panel is a key of the staging colors applied. On the right of the panel are the 45 genes out of CIN70. CIN, chromosome instability; NB, neuroblastoma; MYCN, neuroblastoma MYC oncogene; St., stage.
Moreover, Carter et al (55) produced another CIN selecting the top 25 genes best predicting clinical outcome. Interestingly, we observed that 17/25 genes of the CIN25 showed high expression in HR patients. So that, our preliminary data show that CIN25 gene signature is strongly associated with NB with poor outcome. Moreover, this result indicates that CIN is active and several CIN-related genes are operating in HR-NB.
Finally, we were interested to investigate if upregulated genes could be druggable by Federal Drugs Approved (FDA) approved molecules and we explored the Drug Gene Interaction Database (DGID) (dgidb.genome.wustl.edu/). Interestingly, 3/30 genes upregulated in HR patients stage 4 are targetable by FDA approved drugs (Table I), already used for others disease indicating the possible use of these compounds in NB therapy.
Table I.
Food and drug administration approved drugs targeting chromosome instability-associated genes in neuroblastoma.
| Gene | Drug-gene interaction | PMID references | Disease reports |
|---|---|---|---|
| AURKB | SUNITINIB | 15180525; 14753710; 11752352; 15304385; 12531805; 14654525; 19667267; 27181063; 23392356; 25818407; 17605814; 16685460; 15688612; 17296815; 17367763; 12538485; 12748309; 15557593; 16425993; 24086736; 21478036; 12873999; 17545799; 17046465; 19282169; 25641662; 25592632; 20571495; 18235121; 12181401; 16098458; 15451219; 23177515; 19461405; 18421059; 21642685; 21690468; 22261812; 16638875; 22439647; 18955458; 19861435; 25239608; 23582185; 25594040; 26772734; 19164557; 22357254; 23375402; 21969494; 19723655; 16533791; 16418310; 20142593; 27149458; 28011623; 16648572; 22718859; 19366796; 12949711; 15685537; 15146165; 26130666; 15928335; 18794084; 22745105; 16954519; 14645423; 25157968; 16849418; 20696054; 28447912; 20065189; 22025146; 23056499; 20943719; 21470995; 21422803; 11351254; 20368568; 21455200; 18073307; 19255327; 20847059; 9839497; 9681850; 18541894; 22105611; 26487278; 27751729; 26309414; 24633157; 24606768; 23455880; 26864210 | www.clinicaltrials.gov/ct2/results?cond=&term=SUNITINIB&cntry=&state=&city=&dist=&Search=Search |
| RRM2 | CLADRIBINE | 17852710; 16316309; 19576186; 9923554; 19715446 | www.clinicaltrials.gov/ct2/results?cond=&term=CLADRIBINE&cntry=&state=&city=&dist=&Search=Search |
| GALLIUM NITRATE | 12776257; 1335254; 15651176 | www.clinicaltrials.gov/ct2/results?cond=&term=GALLIUM+NITRATE&cntry=&state=&city=&dist=&Search=Search | |
| GEMCITABINE | None found | www.clinicaltrials.gov/ct2/results?cond=&term=GEMCITABINE&cntry=&state=&city=&dist=&Search=Search | |
| CLOFARABINE | None found | www.clinicaltrials.gov/ct2/results?cond=&term=CLOFARABINE&cntry=&state=&city=&dist=&Search=Search | |
| HYDROXYUREA | None found | www.clinicaltrials.gov/ct2/results?cond=&term=HYDROXYUREA&cntry=&state=&city=&dist=&Search=Search | |
| FLUDARABINE PHOSPHATE | None found | www.clinicaltrials.gov/ct2/results?cond=&term=FLUDARABINE+PHOSPHATE&cntry=&state=&city=&dist=&Search=Search | |
| TOP2A | MITOXANTRONE | 10451375; 11004693; 18687447; 11752352; 9631585; 9494516; 11278845; 9426516 | www.clinicaltrials.gov/ct2/results?cond=&term=CLADRIBINE&cntry=&state=&city=&dist=&Search=Search |
| TENIPOSIDE | 8702194; 16271071; 17361331; 17514873; 11752352; 16480143; 9426516 | www.clinicaltrials.gov/ct2/results?cond=&term=GALLIUM+NITRATE&cntry=&state=&city=&dist=&Search=Search | |
| AMSACRINE | 1322791; 8823806; 10691026; 8519659; 8632768; 11006484; 11716434; 11752352; 11473732; 1311390 | www.clinicaltrials.gov/ct2/results?cond=&term=GEMCITABINE&cntry=&state=&city=&dist=&Search=Search | |
| ETOPOSIDE | 8823806; 9485461; 8870683; 9494516; 9426516 | www.clinicaltrials.gov/ct2/results?cond=&term=CLOFARABINE&cntry=&state=&city=&dist=&Search=Search | |
| PODOFILOX | 16061385; 1334447; 10783066; 11752352; 1845848; 1331331 | www.clinicaltrials.gov/ct2/results?cond=&term=HYDROXYUREA&cntry=&state=&city=&dist=&Search=Search | |
| VALRUBICIN | 11752352; 16019763 | www.clinicaltrials.gov/ct2/results?cond=&term=FLUDARABINE+PHOSPHATE&cntry=&state=&city=&dist=&Search=Search | |
| EPIRUBICIN | 14728934; 16234514; 17639997 | www.clinicaltrials.gov/ct2/results?cond=&term=CLADRIBINE&cntry=&state=&city=&dist=&Search=Search | |
| DOXORUBICIN | None found | www.clinicaltrials.gov/ct2/results?cond=&term=GALLIUM+NITRATE&cntry=&state=&city=&dist=&Search=Search | |
| ENOXACIN | 18471102; 11752352; 10089819 | www.clinicaltrials.gov/ct2/results?cond=&term=GEMCITABINE&cntry=&state=&city=&dist=&Search=Search | |
| DAUNORUBICIN | 9494516 | www.clinicaltrials.gov/ct2/results?cond=&term=CLOFARABINE&cntry=&state=&city=&dist=&Search=Search |
PMID refers to PubMed identifier. All drugs are small molecule drug inhibitor of the respective genes except daunorubicin of which the type of interaction is unknown. The disease report links associated with the pathology lists were treated with the illustrated drug according to the indications derived from www.clinicaltrials.gov/. AURKB, aurora kinase B; RRM2, ribonucleotide reductase regulatory subunit M2; TOP2A, DNA Topoisomerase IIα.
7. Conclusion
CIN is an old genomic aspect that is recently emerged as causative of cancer. Today with the advent of NGS, there is an additional possibility to study in deep this phenomenon. So that, it should be useful to initiate a CIN screening on several cancers which can be exploited for targeted cancer therapy. While numerous studies demonstrated that CIN may promote tumorigenesis, primarily through the functional loss of key players governing chromosome stability, it has also been shown that CIN beyond tolerable levels actually leads to cell death and tumor suppression (56). These observations collectively suggest elevating CIN as a potential chemotherapeutic strategy in a genetically sensitized background, but additional studies would be needed to validate the efficacy and effectiveness of this approach to cancer treatment.
Acknowledgements
The authors would like to thank Dr. Carlo Zanon (Neuroblastoma Laboratory, Fondazione Istituto di Ricerca Pediatrica Città della Speranza, Padua, Italy), for obtaining gene expression profile analysis from the neuroblastoma public dataset.
Funding
The present review was supported by Fondazione Italiana per la Lotta al Neuroblastoma/Fondazione Istituto di Ricerca Pediatrica Città della Speranza (grant no. IRP-13/02).
Availability of data and materials
The datasets used and/or analysed during the current study are included in this published article.
Authors' contributions
GPT conceived the present review. PF and MRE selected the articles and identified useful information within these articles. PF submitted the manuscript. PF, MRE and GPT contributed equally in writing the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Luksch R, Castellani MR, Collini P, De Bernardi B, Conte M, Gambini C, Gandola L, Garaventa A, Biasoni D, Podda M, et al. Neuroblastoma (Peripheral neuroblastic tumours) Crit Rev Oncol Hematol. 2016;107:163–181. doi: 10.1016/j.critrevonc.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Haupt R, Garaventa A, Gambini C, Parodi S, Cangemi G, Casale F, Viscardi E, Bianchi M, Prete A, Jenkner A, et al. Improved survival of children with neuroblastoma between 1979 and 2005: A report of the Italian Neuroblastoma Registry. J Clin Oncol. 2010;28:2331–2338. doi: 10.1200/JCO.2009.24.8351. [DOI] [PubMed] [Google Scholar]
- 3.Defferrari R, Mazzocco K, Ambros IM, Ambros PF, Bedwell C, Beiske K, Bénard J, Berbegall AP, Bown N, Combaret V, et al. Influence of segmental chromosome abnormalities on survival in children over the age of 12 months with unresectable localised peripheral neuroblastic tumours without MYCN amplification. Br J Cancer. 2015;112:290–295. doi: 10.1038/bjc.2014.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tonini GP. Growth, progression and chromosome instability of Neuroblastoma: A new scenario of tumorigenesis? BMC Cancer. 2017;17(20) doi: 10.1186/s12885-016-2986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko Y, Kanda N, Maseki N, Sakurai M, Tsuchida Y, Takeda T, Okabe I, Sakurai M. Different karyotypic patterns in early and advanced stage neuroblastomas. Cancer Res. 1987;47:311–318. [Google Scholar]
- 7.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 8.Stigliani S, Coco S, Moretti S, Oberthuer A, Fischer M, Theissen J, Gallo F, Garavent A, Berthold F, Bonassi S, et al. High genomic instability predicts survival in metastatic high-risk neuroblastoma. Neoplasia. 2012;14:823–832. doi: 10.1593/neo.121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-Z. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 12.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nature Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 13.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. doi: 10.1016/S0962-8924(99)01661-X. [DOI] [PubMed] [Google Scholar]
- 14.Heng HH, Bremer SW, Stevens JB, Horne SD, Liu G, Abdallah BY, Ye KJ, Ye CJ. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013;32:325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 15.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. Aneuploidy vs. gene mutation hypothesis of cancer: Recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci USA. 2000;97:3236–3241. doi: 10.1073/pnas.97.7.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 19.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 20.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 22.Tonini GP. Molecular mechanisms involved in DNA repair, in gene rearrangement and in gene amplification may be considered as an integrated system in maintaining cellular homeostasis and cell survival. Anticancer Res. 1988;8:881–884. [PubMed] [Google Scholar]
- 23.Matzke MA, Mette MF, Kanno T, Matzke AJ. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 2003;19:253–256. doi: 10.1016/S0168-9525(03)00057-X. [DOI] [PubMed] [Google Scholar]
- 24.Gisselsson D. Chromosome instability in cancer: How, when, and why? Adv Cancer Res. 2003;87:1–29. doi: 10.1016/S0065-230X(03)87293-7. [DOI] [PubMed] [Google Scholar]
- 25.Marx J. Debate surges over the origins of genomic defects in cancer. Science. 2002;297:544–546. doi: 10.1126/science.297.5581.544. [DOI] [PubMed] [Google Scholar]
- 26.Sieber OM, Heinimann K, Tomlinson IP. Genomic instability-the engine of tumorigenesis? Nat Rev Cancer. 2003;3:701–708. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 27.Bakhoum SF, Compton DA. Chromosomal instability and cancer: A complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–1143. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 29.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloomfield M, Duesberg P. Inherent variability of cancer-specific aneuploidy generates metastases. Mol Cytogenet. 2016;9:90. doi: 10.1186/s13039-016-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Targa A, Rancati G. Cancer: A CINful evolution. Curr Opin Cell Biol. 2018;52:136–144. doi: 10.1016/j.ceb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Sheltzer JM, Amon A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci USA. 2013;110:E4134–E4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komarova NL, Wodarz D. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc Natl Acad Sci USA. 2004;101:7017–7021. doi: 10.1073/pnas.0401943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera-Rivera Y, Saavedra HI. Centrosome-a promising anti-cancer target. Biologics. 2016;10:167–176. doi: 10.2147/BTT.S87396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cicenas J. The Aurora kinase inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2016;142:1995–2012. doi: 10.1007/s00432-016-2136-1. [DOI] [PubMed] [Google Scholar]
- 38.Kaur R, Kaur G, Gill RK, Soni R, Bariwal J. Recent developments in tubulin polymerization inhibitors: An overview. Eur J Med Chem. 2014;87:89–124. doi: 10.1016/j.ejmech.2014.09.051. [DOI] [PubMed] [Google Scholar]
- 39.Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, Turchi JJ. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharjee S, Nandi S. Synthetic lethality in DNA repair network: A novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life. 2017;69:929–937. doi: 10.1002/iub.1696. [DOI] [PubMed] [Google Scholar]
- 41.Bayani J, Selvarajah S, Maire G, Vukovic B, Al-Romaih K, Zielenska M, Squire JA. Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin Cancer Biol. 2007;17:5–18. doi: 10.1016/j.semcancer.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 43.Bakhoum SF, Kabeche L, Murnane JP, Zaki BI, Compton DA. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 2014;4:1281–1289. doi: 10.1158/2159-8290.CD-14-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heng HH, Spyropoulos B, Moens PB. FISH technology in chromosome and genome research. BioEssays. 1997;19:75–84. doi: 10.1002/bies.950190112. [DOI] [PubMed] [Google Scholar]
- 45.Imle A, Polzer B, Alexander S, Klein CA, Friedl P. Genomic instability of micronucleated cells revealed by single-cell comparative genomic hybridization. Cytometry A. 2009;75:562–568. doi: 10.1002/cyto.a.20733. [DOI] [PubMed] [Google Scholar]
- 46.Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, Alaiya A, Sennerstam R, Zetterberg A, Auer G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–909. doi: 10.1158/0008-5472.CAN-03-2451. [DOI] [PubMed] [Google Scholar]
- 47.Mullauer L. Next generation sequencing: Clinical applications in solid tumours. Memo. 2017;10:244–247. doi: 10.1007/s12254-017-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 50.Schukken KM, Foijer F. CIN and aneuploidy: Different concepts, different consequences. Bioessays. 2018;40 doi: 10.1002/bies.201700147. [DOI] [PubMed] [Google Scholar]
- 51.Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A, Valdora F, Fischer M, Gallo F, Hero B, et al. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer. 2012;131:1591–1600. doi: 10.1002/ijc.27432. [DOI] [PubMed] [Google Scholar]
- 52.Masecchia S, Coco S, Barla A, Verri A, Tonini GP. Genome instability model of metastatic neuroblastoma tumorigenesis by a dictionary learning algorithm. BMC Med Genomics. 2015;8:57. doi: 10.1186/s12920-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schleiermacher G, Janoueix-Lerosey I, Delattre O. Recent insights into the biology of neuroblastoma. Int J Cancer. 2014;135:2249–2261. doi: 10.1002/ijc.29077. [DOI] [PubMed] [Google Scholar]
- 54.Schnepp RW, Khurana P, Attiyeh EF, Raman P, Chodosh SE, Oldridge DA, Gagliardi ME, Conkrite KL, Asgharzadeh S, Seeger RC, et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28:599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 56.Chan SH, Ngeow J. Germline mutation contribution to chromosomal instability. Endocr Relat Cancer. 2017;24:T33–T46. doi: 10.1530/ERC-17-0062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are included in this published article.