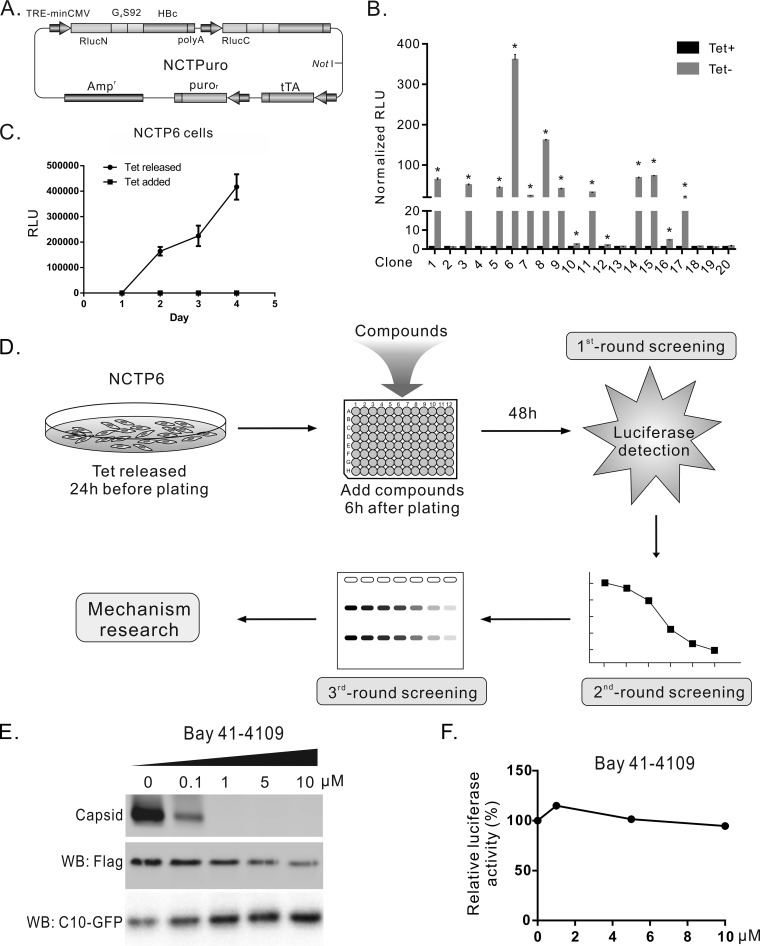
FIG 3.
Establishment of stable cell line SRluc-HBc6 and compound screening procedure. (A) Schematic illustration of plasmid NCTPuro. The plasmid contains 4 open reading frames, which express an RlucN-HBc, an RlucC-HBc, a puromycin resistance gene, and a tetracycline-controlled transactivator (tTA). (B) Selection and characterization of stable cell lines. Twenty clones were obtained after puromycin selection. Rluc activities of these clones were detected in the presence and absence of tetracycline. (C) Time course measurement of Rluc activity in SRluc-HBc6 cells. (D) The diagram of the compound screening procedure. SRluc-HBc6 cells cultured in a 10-cm diameter dish were seeded into 96-well plates with the tetracycline-free medium. Six hours after seeding, compounds were added, and Rluc activity was assayed 48 h after treatment for the first-round screening. Compounds identified from the first-round screening were tested for dose-effect curves in SRluc-HBc6 cells as the second-round screening. For the third-round screening, compounds were tested in HepAD38 cells for their effects on HBV DNA replication by Southern blotting. (E) Bay 41-4109 inhibited capsid formation. HepG2 cells cotransfected with 3×Flag-G4S92-HBc and C10-GFP were treated with Bay 41-4109 for 6 days. Intracellular capsids were detected by particle gel assay. The expression of the recombinant core protein and C10-GFP (control) was detected by Western blotting. (F) Bay 41-4109 did not affect the Rluc activity of SRluc-HBc6 cells. SRluc-HBc6 cells were treated for 2 days with Bay 41-4109 of different concentrations, and the Rluc activity was assayed.