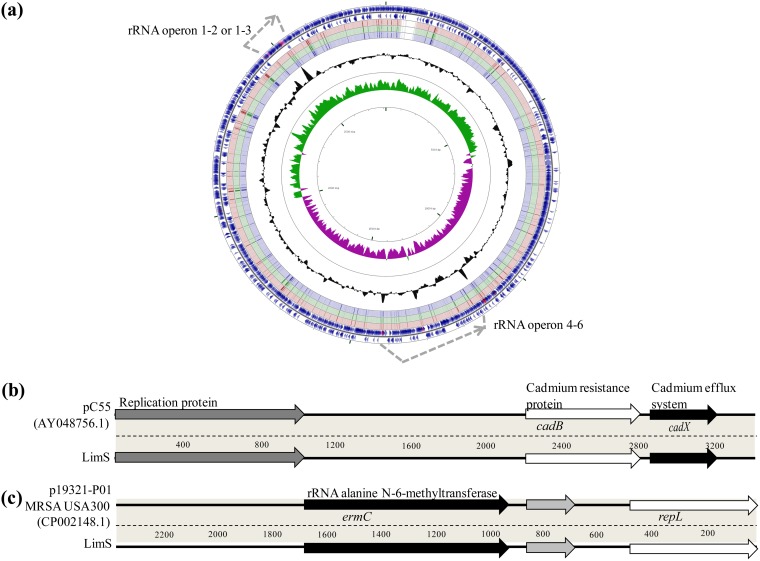
FIG 2.
Genome features of the chromosome (a) and the 2 plasmids (b and c) identified in Lim isolates. (a) Comparison by BLAST of the coding sequences (CDS) of the FORC_012 (blue arrows of the 2 outer rings) and LimS (red), LimR1 (green), and LimR12 (purple) genomes (corresponding to the 3rd, 4th, and 5th rings, respectively, from outside to inside) (CGview server [56]). GC content is represented on the plot inside the ring. The major difference between the FORC_012 genome and those of Lim isolates is the number of rRNA operons (gray dotted-line arrows), with 6 rrl genes for FORC_012 conversely to 5 for Lim isolates, corresponding to the loss of the 3rd operon. The other difference is the presence of a supplementary prophage in the FORC_012 genome. (b and c) Genes are labeled on the coding sequences for the 2 small cryptic plasmids of isolate LimS.