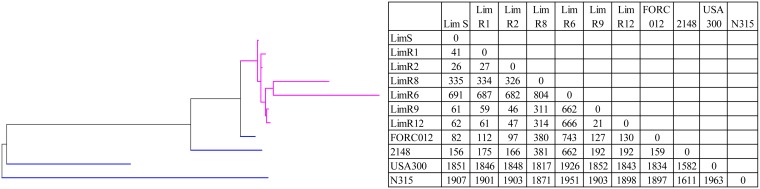
FIG 4.
Results of phylogenetic analysis of the 7 MRSA isolates analyzed by wgMLST. A maximum parsimony tree (Bionumerics) with default parameters was used for the analysis of all loci (wgMLST and MLST alleles). The root position in the tree was assigned to the deepest branch measured by average branch length. S. aureus strains FORC_012 (ST72, t664), 2148 (ST72), N315 (ST5), and USA300-FPR3757 (ST8) were used for reference genomes (blue). Lim isolate branches are depicted in purple. Numbers in the body of the table represent the pairwise wgMLST allelic differences between isolates.