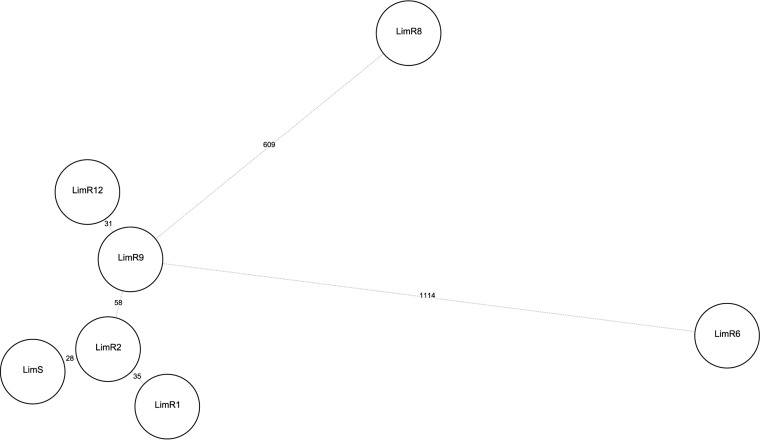
FIG 5.
Evolution of the pairwise distances (SNPs) in comparison with the LZD-susceptible Lim isolate, LimS, based on SNP analysis. A minimum spanning tree (BioNumerics version 7.6) with default parameters (priority rule1, maximum number of N locus variants [N = 1]; weight, 10,000) was used. Numbers on the tree branches indicate SNP distances between genomes (circles). Genome codes refer to Lim strain numbers, which are chronologically ordered as shown in Fig. 1.