Abstract
Epigenetic drift and age-related methylation have both been used in the literature to describe changes in DNA methylation that occurs with aging. However, ambiguity remains regarding the exact definition of both of these terms, and neither of these fields of study explicitly considers the impact of environmental factors on the aging epigenome. Recent twin studies have demonstrated longitudinal, pair-specific discordance in DNA methylation patterns, suggesting an effect of the environment on age-related methylation and/or epigenetic drift. Supporting this idea, other new reports have shown clear environment- and toxicant-mediated shifts away from the baseline rates of age-related methylation and epigenetic drift within an organism, a process we now term “environmental deflection.” By defining and delineating environmental deflection, this contemporary review aims to highlight the effects of specific toxicological factors on the rate of DNA methylation changes that occur over the life course. In an effort to inform future epigenetics-based toxicology studies, a field of research now classified as toxicoepigenetics, we provide clear definitions and examples of “epigenetic drift” and “age-related methylation,” summarize the recent evidence for environmental deflection of the aging epigenome, and discuss the potential functional effects of environmental deflection.
Keywords: exposure, environmental (Environmental Toxicology, aging), (Reproductive & Developmental Toxicology, developmental toxicity), prenatal (Reproductive & Developmental Toxicology, developmental toxicity), post-natal (Reproductive & Developmental Toxicology).
Increasing evidence supports the developmental origins of health and disease (DOHaD) paradigm, which posits that exposure to environmental factors (eg, diet, chemicals, stress, etc.) during critical periods of life (eg, pre-conception, gestation, infancy, adolescence) alters disease susceptibility later in life by influencing developmental plasticity (Bateson et al., 2004, Heindel et al., 2015). As support for DOHaD accumulates, it has been shown that developmental exposure to environmental factors can alter gene regulation and subsequent phenotype through changes in the epigenome (Waterland and Jirtle, 2004; Waterland and Michels, 2007); this field of research is now classified as “toxicoepigenetics.”
Epigenetics refers to the study of heritable and potentially reversible changes in gene expression unrelated to the DNA sequence. Epigenetic marks include chromatin remodeling modifications (eg, histone tail trimethylation), non-coding RNA, and alterations to DNA itself (eg, DNA methylation) (Egger et al., 2004; Bernal and Jirtle, 2010). DNA methylation is a well-characterized epigenetic control mechanism, and is typically defined as the addition of a methyl group to the 5’-carbon of cytosine in a Cytosine-phospho-Guanine (CpG) dinucleotide— 5-methylcytosine (5-mC). In general, DNA methylation is associated with decreased transcription factor binding at promoter/enhancer sites, as well as decreased gene transcription (Medvedeva et al., 2014). Previous work have documented distinct waves of demethylation and de novo methylation that occur during fetal development (Reik et al., 2001; Smallwood and Kelsey, 2012), as well as evidence that these waves of epigenetic reprogramming help regulate primordial germ cell proliferation and differentiation (Messerschmidt et al., 2014).
Although epigenetic reprogramming events are typically tightly regulated, 5-mC levels have been shown to change in response to environmental exposures during early development (Bernal and Jirtle 2010; Anderson et al., 2012; Manikkam et al., 2013), adolescence (Essex et al., 2013), and even adulthood (Wright et al., 2010; Tellez-Plaza et al., 2014). For example, animal studies have demonstrated that offspring DNA methylation is associated with developmental exposure to a variety of environmental factors, including lead (Pb) (Dosunmu et al., 2012), altered diet (Vucetic et al., 2010, Marco et al., 2014), vinclozolin (Guerrero-Bosagna et al., 2012), arsenic (Reichard and Puga 2010), bisphenol A (BPA) (Kim et al., 2014), trichloroethylene (TCE) (Gilbert et al., 2012), ethanol (EtOH)(Kaminen-Ahola et al., 2010; Laufer et al., 2013; Marjonen et al., 2015), diesel exhaust (DE) (Tachibana et al., 2015), and stress (Dong et al., 2015). Although this review focuses mainly on DNA methylation, there is also evidence that environmental factors may influence other epigenetic modifications including posttranslational histone tail modifications (Arita et al., 2012), overall chromatin state (Schick et al., 2015; Veazey et al., 2015), and DNA hydroxymethylation (Tammen et al., 2014).
AGE-RELATED METHYLATION AND EPIGENETIC DRIFT: TWO TYPES OF CHANGE OVER THE LIFE COURSE
Although there has been a heavy focus in the environmental health sciences literature on the association between toxicological factors and the epigenome in cross-sectional studies and in utero exposure models, a number of molecular epidemiology and genomics studies have evaluated DNA methylation status as a function of age in humans and animal models. The aging epigenome was first described thirty years ago, when early investigations showed that levels of CpG methylation in human fibroblast cells and pooled mouse tissues were inversely related to lifespan (Fairweather et al., 1987; Wilson et al., 1987). More recently, a large number of studies have demonstrated age-dependent changes in DNA methylation, including twin studies (Fraga et al., 2005; Martino et al., 2013), human cohort studies (Alisch et al., 2012; Heyn et al., 2012; Madrigano et al., 2012; Wang et al., 2012; Urdinguio et al., 2016), and animal model studies (Maegawa et al., 2010; Spiers et al., 2016). Among the classically defined epigenetic marks, DNA methylation is most often investigated in epigenetic aging studies because of its stability and the availability of high throughput quantification methods. Studies investigating the aging epigenome show some consistent patterns, including locus-specific increases in DNA methylation with age (Teschendorff et al., 2013), global decreases in DNA methylation with age (Teschendorff et al., 2013; Issa, 2014), and bidirectional changes in DNA methylation variability over time (Shah et al., 2014; Jones et al., 2015). To describe the epigenomic changes that occur in conjunction with chronological age, the literature has settled on two terms—age-related methylation and epigenetic drift (Issa, 2014; Jung and Pfeifer, 2015). Here, we aim to define and distinguish age-related methylation and epigenetic drift, and then later describe environmental deflection as the effects of environmental exposures on both phenomena.
Age-related methylation is traditionally defined as predictable, direction-specific changes in DNA methylation levels that occur with normal aging (Jung and Pfeifer, 2015). This concept is closely linked to the “epigenetic clock” proposed by Horvath, which showed that biological age could be reliably predicted from DNA methylation levels at specific CpG sites across the genome (Horvath, 2013). Results from the literature demonstrate that age-related methylation occurs both at specific gene regions (Jung and Pfeifer, 2015) and on an epigenome-wide scale (Heyn et al., 2012). Additionally, a recent review of the aging epigenome noted that the directionality of age-related methylation—hypomethylation or hypermethylation—varies by gene region (Jones et al., 2015). Considered together, these results suggest that age-related methylation is a complex process that can vary by genomic context. Further supporting this idea, age-related methylation has been shown to vary by tissue type. Day et al. looked at methylation array data from four different human tissue types—blood, kidney, brain, and skeletal muscle—and found both tissue-independent and tissue-dependent methylation changes associated with age (Day et al., 2013). Based on the age-related methylation changes common to multiple tissue types, the authors assert that age-related methylation is not stochastic, and may be biologically meaningful.
In contrast to age-related methylation, epigenetic drift refers to stochastic, bidirectional changes in epigenetic (eg, DNA methylation) variability with age (Jones et al., 2015). These changes, which may alter methylome plasticity, are thought to be a result of methylation maintenance failure during cellular replication (Fraga et al., 2005; Teschendorff et al., 2013). Unlike an age-related methylation, epigenetic drift is not a predictable process; instead, it can be conceptualized as the direct result of random inefficiencies in biological machinery that occur with age (Shah et al., 2014; Jones et al., 2015). As such, epigenetic drift is not expected to be consistent across individuals within a population, and cannot be used to the predict age. Nevertheless, this concept is critical for describing the epigenetic discordance that arises in monozygotic twins as they age (Fraga et al., 2005), and can also help explain results from cross-sectional studies that show increased epigenetic variability with advanced age (Talens et al., 2012). Baseline levels of epigenetic drift are expected to occur regardless of specific environmental exposures, providing a background rate of increased variability that occurs in tandem with site-specific age-related methylation changes. Supporting this idea, a recent study found an interaction between epigenetic drift and age-related methylation at specific “epigenetic clock” CpG sites, showing an increased effect of environmental or stochastic influences with increasing age (van Dongen et al., 2016). This suggests that the relative contribution of epigenetic drift to the aging epigenome (ie, longitudinal DNA methylation) varies across the lifecourse.
ENVIRONMENTAL DEFLECTION OF THE AGING EPIGENOME
Studies suggest that both age-related methylation and epigenetic drift are affected by exposure to environmental factors. For example, a recent twin cohort study demonstrated longitudinal, pair-specific DNA methylation divergence, indicating a possible interaction between the environment and age-related methylation (Martino et al., 2013). Of particular interest to the field of toxicology, additional reports indicate that environmental exposure to exogenous environmental factors (eg, lead (Pb), altered diet) can alter the rate of either age-related methylation (Faulk et al., 2014; Kochmanski et al., 2016) or epigenetic drift (Gilbert et al., 2016) across the life course. These results indicate that an interaction between age and exposure exists, and that investigations into the effects of environmental exposure on DNA methylation should not be limited to cross-sectional analyses. However, although these papers discuss a role of the environment in establishing rates of “epigenetic drift” and/or “age-related methylation,” they do not provide a specific mechanism by which the environment could shape the aging epigenome.
In an effort to improve clarity and interpretation of epigenetic studies in both animal models and human cohorts, we propose a new term for this mechanism—environmental deflection—that refers to an environment- or toxicant-mediated shift away from the baseline rate of age-related methylation or epigenetic drift within an organism. By altering longitudinal patterns of epigenetic marks, environmental deflection may facilitate long-term changes in gene regulation via specific environmental exposures, showing the greatest effects during critical periods of growth and development. As such, environmental deflection may underlie the apparent delay between developmental exposure and biological effects later in life. This type of long-lived, toxicant-sensitive epigenetic mechanism may also help to explain the growing prevalence of chronic diseases in human populations, demonstrating that longitudinal measures of the epigenome should be considered when designing future toxicoepigenetic and epigenetic epidemiological studies.
Conceptual Model for Environmental Deflection
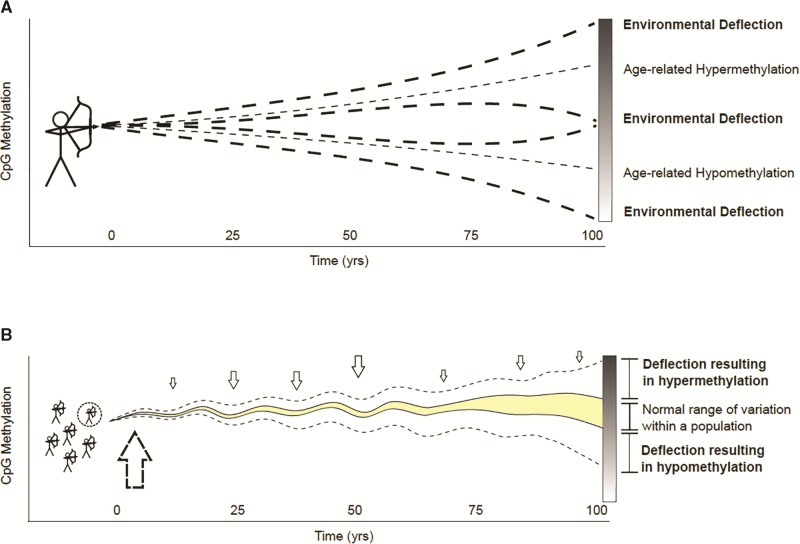
In an effort to establish a clear conceptual framework for environmental deflection, we have utilized a visual bow and arrow metaphor that takes into account exposure timing and age-related methylation (Figure 1) . Time zero in our model represents the period of initial developmental epigenetic programming, the flight of the arrow represents the rate of age-related methylation, and deviations in the flight path represent environmental deflection. Figure 1A demonstrates how epigenetic status at a specific locus or global marker can change with age or be deflected by an environmental exposure away from age-related hyper- or hypomethylation. Respectively, these endpoints can be conceptualized as the arrow striking its target and the arrow missing the target by a significant amount. Figure 1B shows the environmental deflection of epigenetic drift for a population; the shaded area is the normal range of DNA methylation variability for the population. If a subset of the population (dotted circle) is exposed to specific toxicant exposures (vertical dotted arrows) during the early developmental time period (large vertical dotted arrow), the trajectory over time at labile genes could be deflected outside the normal range of variation, depicted by the dotted lines. When firing a real-life arrow, the greatest opportunity to affect the arrow’s flight occurs at release. Assuming environmental deflection works in a similar way—as suggested by developmental plasticity theory (Wells, 2014)—the greatest opportunity to affect the normal trajectory of age-related methylation and/or epigenetic drift (ie, the arrow’s flight) is early in life (ie, the release point). However, much like wind or other outside force can alter the path of an arrow after release, environmental deflection can also occur at other points throughout an organism’s life course (Figure 1B). To test this conceptual model in real populations, longitudinal studies with adequate early life exposure data and repeated epigenetic assessments are recommended to identify epigenetic loci with deflected methylation measurements.
FIG. 1.
A conceptual framework for environmental deflection of the aging epigenome. (A) Environmental deflection of age-related methylation for an individual; deflection is represented by the altered flight of an arrow fired at a target. The gradient bar on the right shows how epigenetic status at a specific locus or global marker can change with age or be deflected by an environmental exposure away from age-related hyper- or hypomethylation. Respectively, these endpoints can be conceptualized as the arrow striking its target and the arrow missing the target by a significant amount. (B) Environmental deflection of epigenetic drift for a population; the vertical arrows represent specific exposures that may affect drift trajectory throughout life. The shaded area is the normal range of DNA methylation variability at a given time point for the population. If a subset of the population (dotted circle) is exposed to a toxicant during the early developmental time period (large vertical dotted arrow), the epigenetic drift trajectory at labile genes could be deflected outside the normal range of variation, depicted by the dotted line.
EVIDENCE FOR ENVIRONMENTAL DEFLECTION
Epigenetic Discordance and Variability in Human Twin Studies
As a byproduct of their identical genetic background, monozygotic twin pairs are ideal for investigating the role the environment plays in shaping the aging epigenome while controlling for genetic effects. In 2005, a landmark paper by Fraga et al. demonstrated divergence of DNA methylation status with age in separate identical twin populations (Fraga et al., 2005). Although this study was not longitudinal, the results suggest that the environment and lifestyle, not simply genetics, is driving age-associated changes in human methylation status. A more recent report demonstrated that newborn monozygotic (MZ) twins exhibit distinct patterns of inter-individual DNA methylation variation, indicating that the environment plays a role in determining the neonatal methylome (Ollikainen et al., 2010). Building off these ideas, Talens et al. found locus-specific increases in within-twin pair methylation discordance across the adult life course (18-49 years old), a pattern that was attributable to the individual’s unique environment at most investigated loci (Talens et al., 2012). Extending twin studies to an epigenome-wide scale, several newer studies have investigated age-related changes in DNA methylation in twin pairs using Illumina BeadChips (Bell et al., 2012; Lévesque et al., 2014; van Dongen et al., 2016). These studies have shown region-specific hypermethylation with age in adult MZ twins (Bell et al., 2012), high levels of within-pair DNA methylation variability in adolescent twins (Lévesque et al., 2014), and significant interaction between environmental effects and age at 32,234 CpG sites across the epigenome (van Dongen et al., 2016). Combined, the results from these twin studies support the idea that the environment plays an integral role in shaping the epigenome throughout human aging. However, by their very nature, twin studies are not able to tease apart the separate effects of environmental factors on epigenetic drift and age-related methylation, and do not examine interactions between specific exposures and the aging epigenome.
Human Environmental Deflection Studies
Supplementing the available twin studies, a handful of recent non-twin human cohort studies have examined environmental deflection of epigenetic drift or age-related methylation by toxicants and other lifestyle factors. Building off the smaller scale methylation array and global methylation results noted previously, a recent publication used the Illumina 450K BeadChip to examine the effects of exposure on epigenome-wide DNA methylation at >450,000 CpG sites in blood samples from a human cohort (Shah et al., 2014). Within this study, Shah et al. showed that smoking-associated CpG probes exhibited diminished epigenetic drift, suggesting an environmental deflection of the drift rate by smoking status. Similarly, a recent publication from Horvath et al. showed that increased body mass index (BMI) was associated with accelerated age-related methylation at 353 CpG sites in human liver samples, suggesting that nutritionally-induced oxidative stress and metabolic alterations may deflect the rate of age-related methylation (Horvath et al., 2014). These results indicate that environmental deflection of both epigenetic drift and age-related methylation may occur on an epigenome-wide scale in human populations.
Although specific exposures can be difficult to quantify in human populations, several studies have evaluated stress events as definable representations of the human environment, examining environmental deflection of age-related methylation by stress (Boks et al., 2015; Zannas et al., 2015; Brody et al., 2016). The first of these papers, Boks et al. 2015, found that traumatic stress in soldiers deployed in Afghanistan significantly accelerated age-related DNA methylation in blood samples, suggesting a stress-mediated modification to the rate of epigenetic aging, and evidence that environmental deflection is not strictly limited to a developmental time period (Boks et al., 2015). Complementing this result, Zannas et al. demonstrated that cumulative lifetime stress was associated with an increased rate of age-related DNA methylation in blood (Zannas et al., 2015), and a 2016 study by Brody et al. showed that harsh parenting during childhood also had an effect on age-related methylation in blood samples (Brody et al., 2016). Considered together, these recent human studies indicate that stress exposures at various life course stages are associated with modifications to age-related methylaton in human cohorts, supporting a model of environmental deflection that is dynamic throughout life (Figure 1B).
Animal Model Environmental Deflection Studies
Although it is difficult to examine the effects of single exposures in human populations, a number of animal experiments have indicated that prenatal toxicant exposures (eg, lead, ethanol, high-fat diet, etc.) alter the establishment of DNA methylation marks during development, and several reports have shown that these environment-induced changes in DNA methylation may be carried over into adulthood (Dosunmu et al., 2012; Laufer et al., 2013; Marco et al., 2014). However, most of the animal studies investigating the effect of environmental factors on DNA methylation utilize cross-sectional CpG methylation as an outcome, and are thereby unable to answer an important question: do prenatal environmental exposures modify the rates of age-related methylation or epigenetic drift?
A small number of recent animal model studies have begun to address this question, showing environmental deflection by exposure to experimentally controlled environmental factors. For example, using paired mouse tail tissue, Faulk et al. found that perinatal exposures to lead (Pb) altered the rate of age-related methylation in the promoter of a murine imprinted gene, Igf2r, as well as a metastable epiallele, CabpIAP (Faulk et al., 2014). Similarly, using paired mouse tail tissue, we demonstrated that perinatal exposure to high-fat diet (HFD) altered the rate of age-related hypomethylation at conserved CpG sites within the Intracisternal A-Particle (IAP) class of retrotransposons (Kochmanski et al., 2016). Comparing tail DNA methylation from postnatal day 21 (PND21) and 10 months of age, we found a steeper rate of age-related IAP hypomethylation in mice exposed to HFD compared to control. Additionally, HFD-exposed mice showed a significant increase in PND21 methylation at the Igf2 imprinted locus compared to control, suggesting a pattern of exposure-induced premature epigenetic aging at this particular gene region. Examining epigenetic drift, a third study in mice tested whether adult trichloroethylene (TCE) exposure would alter DNA methylation variance in CD4+ T cells across the time-course of exposure (40 weeks) (Gilbert et al., 2015). For this study, the authors used bisulfite sequencing to compare longitudinal CD4+ T cell methylation at specific gene regions between TCE-exposed and control mice. They demonstrated a TCE-dependent increase in naïve CD4+ T cell methylation variance at several gene regions, indicating that epigenetic drift can be shifted away from baseline by a chemical exposure. Together, these animal studies provide direct, exposure-specific evidence for environmental deflection of both age-related methylation and epigenetic drift.
THEORETICAL EXAMPLES OF AGE-RELATED METHYLATION, EPIGENETIC DRIFT, AND ENVIRONMENTAL DEFLECTION
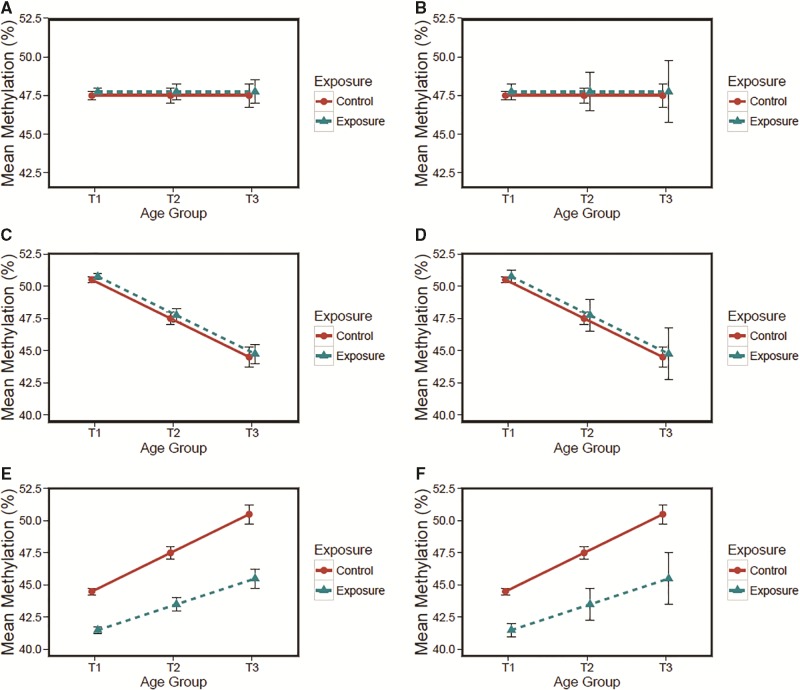
The recent human and animal studies reviewed above indicate that exposure to defined environmental factors can deflect the rates of both epigenetic drift and age-related methylation. To further clarify potential interactions between age-related methylation, epigenetic drift, and environmental deflection, we have generated a collection of theoretical aging epigenome scenarios (Figure 2). Figure 2 presents average DNA methylation and standard error from a single tissue type for a hypothetical study population sampled at three time points (T1 = birth, T2 = adolescence, and T3 = middle-age). Although not exhaustive, a number of possible scenarios are included: A. no age-related methylation, baseline epigenetic drift (Figure 2A); B. no age-related methylation, environmental deflection of epigenetic drift (Figure 2B); C. normal age-related hypomethylation, baseline epigenetic drift (Figure 2C); D. normal age-related hypomethylation, environmental deflection of epigenetic drift (Figure 2D); E. environmental deflection of age-related hypermethylation, baseline epigenetic drift (Figure 2E); F. environmental deflection of both age-related hypermethylation and epigenetic drift (Figure 2F). It is likely that the perfect linear relationships presented in our theoretical figures will not carry over to data collected from real tissue, but Figure 2 does provide a visual representation for the concepts of age-related methylation, epigenetic drift, and environmental deflection, and further indicates the potential interactions between all three concepts. Future studies are needed to identify whether the specific genetic loci exhibit the patterns indicated in our theoretical examples.
FIG. 2.
Theoretical aging epigenome scenarios. Average DNA methylation and standard error at one CpG site from a single tissue type are shown for a hypothetical study population sampled at three time points (T1= birth, T2 = adolescence, and T3 = middle-age). Possible scenarios include but are not limited to: (A) no age-related methylation, baseline epigenetic drift, (B) no age-related methylation, environmental deflection of epigenetic drift, (C) normal age-related hypomethylation, baseline epigenetic drift, (D) age-related hypomethylation and environmental deflection of epigenetic drift, (E) environmental deflection of age-related hypermethylation, and baseline epigenetic drift, and (F) environmental deflection of both age-related hypermethylation and epigenetic drift. Changes could have occurred gradually over time or suddenly at a specific developmental time point.
MECHANISMS AND PHENOTYPIC CONSEQUENCES OF ENVIRONMENTAL DEFLECTION
Mechanisms
To understand how environmental exposures influence epigenetic drift or age-related methylation, we must first consider the mechanisms that underlie life course changes. Current theory holds that epigenetic drift is largely driven by the accumulation of errors in epigenetic maintenance machinery, leading to gradual loss of methylation in hypermethylated regions and gain in hypomethylated regions. Supporting this theory, higher rates of epigenetic drift are observed in proliferative tissues (Day et al., 2013; Issa, 2014; Oh et al., 2016). In addition to random errors, chromatin structure and protein-DNA interactions may influence the probability of epigenetic drift occurring in a given genomic region. DNA methyltransferase 1 (DNMT1) often occupies hypermethylated genes bodies and bivalent chromatin. The loss of DNMT1 binding can lead to a breakdown in chromatin boundaries over time, thereby facilitating the spread of methylation into gene promoters and loss of methylation in gene bodies (Day et al., 2013). Separate from DNMT1 binding effects, regions of the epigenome that are critical for health may be targeted for epimutation repair or protected from errors indirectly by gene expression machinery (Issa, 2014). For example, the Sp1 and Sp3 transcription factors, as well as RNA Polymerase II, are associated with resistance to de novo methylation in promoter regions (Boumber et al., 2008; Takeshima et al., 2009). Similar DNA-protein interactions likely influence age-related methylation and epigenetic drift.
Additional insights into potential mechanisms can be gleaned from research that examines the relationship between cancer and accelerated epigenetic aging. Research shows that steroid receptor mutations in breast cancer are associated with accelerated epigenetic aging, suggesting that changing steroid hormone levels or response across the life course may influence epigenetic aging in some tissues (Horvath, 2013). Although hypermethylation of the promoters of polycomb group protein target genes is observed in both cancer and age-related methylation (Teschendorff et al., 2010), the regulation of genes such as Ezh2, shown to prevent this response, across the life course may influence when hypermethylation occurs (Hasegawa et al., 2016). As such, it is possible that steroid hormone levels or epigenetic regulators—ie, polycomb group proteins—are involved in environmental deflection of the aging epigenome.
Given the many players involved in epigenetic drift and age-related methylation, one can envision multiple avenues of disruption by environmental toxicants that would alter rates of epigenetic drift and/or aging. Epigenetic drift or age-related methylation could be accelerated by exposures that directly inhibit epigenetic machinery (eg, DNMT1, TET1) or indirectly alter available levels through changes in signaling and gene expression. For example, developmental Pb exposure is associated with decreased expression of DNA methyltransferases and methyl-binding protein in adult monkeys (Bihaqi et al., 2011). Alternatively, environmental deflection may be mediated by an inflammatory response or oxidative stress, biological processes that are induced by toxicant exposure and may alter the epigenome. Although inflammation is hypothesized to influence the aging epigenetic profile by inducing stem cell proliferation (Issa, 2014), oxidative stress activates TET and impacts the pool of methyl donors available through the one-carbon metabolism pathway (Kalani et al., 2014; Chia et al., 2011). It is likely that all of these factors contribute to environmental deflection, but additional research is needed to elucidate critical periods of susceptibility and the mechanisms underlying deflection by specific toxicants.
Phenotypic Consequences
The potential phenotypic implications of environmental deflection are evident in monozygotic twins discordant for disease. Although monozygotic twins share genetics and a portion of the early-life environment, they exhibit discordance for a number of diseases—cancers, schizophrenia, diabetes, and autism spectrum disorders (ASD) (Castillo-Fernandez et al., 2014). Concordant with disease, both environmental exposures and the epigenome of monozygotic twins diverge with age (Fraga et al., 2005), suggesting that environmental deflection may play a role in twin disease discordance. Epigenetic differences associated with disease status have been discovered in twin studies for an array of diseases including type I and II diabetes, schizophrenia, and cancers (Castillo-Fernandez et al., 2014). As an example, one study showed differentially methylated sites (DMS), including genetically independent DMS, by type II diabetes status among monozygotic twins (ages >40 years). DMS were enriched in disease-relevant pathways (eg, insulin sensitivity), and some were replicated in a cohort of unrelated cases and controls (Yuan et al., 2014). As a second example, a study of 15-year old monozygotic twins found DMS associated with Autism Spectrum Disorder (ASD) or ASD-related behaviors in blood leukocyte DNA (Wong et al., 2014). However, due to the cross-sectional nature of these studies, we cannot determine whether the DMS were a result of toxicant-mediated environmental deflection, stochastic differences in the intrauterine environment experienced by each twin, or the disease phenotype itself. To provide better evidence for environmentally-labile regions of the epigenome that change over time and contribute to disease onset, future studies could interrogate the epigenome longitudinally via archived samples (eg, neonatal bloodspots) and matched samples at recruitment.
Further insight into the phenotypic implications of environmental deflection can be gained from the literature on the aging epigenome. In general, past studies suggest that the aging epigenome is characterized by hypomethylation of repetitive elements and hypermethylation of specific regions (eg, CpG islands), patterns also observed in many cancerous tissues (Teschendorff et al., 2013; Yuan et al., 2015). Some age-related hypermethylation serves a clear biological purpose—for example, shutting down developmental genes (Teschendorff et al., 2013). On the other hand, age-related changes in methylation are also enriched in pathways involved in stem cell differentiation (West et al., 2013) and may lead to epigenetic mosaicism within stem cells, a process linked to improper, selective cell proliferation (Issa, 2014). These changes in the stem cell population can increase risk for cancers and other disorders more common among the elderly, suggesting a mechanism by which environmental deflection of the aging epigenome could alter predisposition for chronic disease development. Accelerated epigenetic age has recently been associated with all-cause mortality, cancer mortality, and Alzheimer’s Disease neuropathology and memory decline (Levine et al., 2015; Chen et al., 2016; Perna et al., 2016). Environmental exposures that speed up age-related methylation and/or epigenetic drift could increase risk of disease earlier in the life course; alternatively, protective exposures could decrease epigenetic aging at key loci (Figure 2E and F). Based on this proposed mechanism, a better understanding of environmental deflection patterns could assist in the development of interventions that decrease risk for chronic disease.
FUTURE DIRECTIONS
Thorough characterization of epigenetic drift, age-related methylation, and environmental deflection could help inform intervention strategies, identify and protect sensitive populations, and improve later-life disease outcomes. However, this will only be possible with thoughtful study design and sample collection/storage methods that consider the following limitations.
Distinguishing between expected age-related change and exposure-mediated deflection in humans may be difficult for a number of reasons, including limited level of variable control, inherent cohort bias, and/or missing exposure data. These limitations could introduce confounding factors that must be accounted for during data analysis. In addition to potential confounding, much of the human aging epigenome literature utilizes DNA sourced from advanced age cohorts, a study design that presents two major design issues: first, it is difficult to assess the effects of early-life exposure on long-term epigenetic changes; second, it is not possible to determine whether age-related diseases are the cause or the effect of measured epigenetic changes. Recent evidence shows a pattern of increased DNA methylation early in life followed by a gradual loss in adulthood (Jones et al., 2015); however, there are very few human studies that have co-investigated age-related epigenetics and exposure effects in the context of early-life. As such, it is difficult to pinpoint exact periods of exposure susceptibility when discussing longitudinal DNA methylation patterns. Research also suggests a convergence of DNA methylation patterns at the very late stages of human life (Oh et al., 2016), indicating that epigenetic patterns measured from advanced age cohorts may differ from those that occur during adolescence or middle-age, when many chronic diseases begin to develop. In addition to utilizing samples from individuals of advanced age, many human cohort epigenetic studies focus on a single target tissue—peripheral blood leukocytes. Although bioavailable tissues are a necessity for human studies, a recent paper from Day et al. showed that age-related patterns in DNA methylation are tissue-specific (Day et al., 2013), suggesting that age-related epigenetic patterns in a single tissue may not reflect the entire organism. This complicates the interpretation of human epigenetic data, highlighting the importance of integrating results from stem cell and animal model studies.
Longitudinal animal exposure studies provide large sample sizes, a controlled developmental environment, controlled genetics (if using inbred strains or genetically diverse strains such as the collaborative cross), and a number of available tissue types, making them ideal for investigating environmental deflection. Despite these strengths, relatively few longitudinal animal model experiments have examined the effects of environment exposures on life course epigenetics. To gain a true understanding of the dynamics between environmental toxicants, aging, and the epigenome, the field of toxicoepigenetics must shift the focus from cross-sectional exposure studies to animal studies that investigate the relationship between developmental toxicant exposure, longitudinal DNA methylation, and phenotypic information on disease outcomes. The Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET II) program, funded by the US National Institutes of Environmental Health Sciences (NIEHS), is a current research consortium that will evaluate environmental deflection by several toxicant exposures in multiple murine tissue types. These types of future studies should consider environmental deflection of both age-related methylation and epigenetic drift, simultaneously measuring the effects of exposure on longitudinal changes in absolute methylation and methylation variance (Figure 2F). Taken together, this would allow for a comprehensive investigation of environmental deflection as a mediator of exposure-induced disease.
Although there are a number of advantages to animal model studies, they remain expensive, time-consuming, and bring with them a host of ethical concerns. In contrast to animal studies, human stem cells provide an in vitro method for capturing epigenome dynamics during cellular aging. A recent review of research investigating stem cells and aging indicates that the epigenome of human stem cells changes with age, showing patterns of hypo/hyper-methylation that may affect stem cell function (Goodell and Rando, 2015). As a specific example, recent research showed a bimodal pattern of age-dependent epigenome dynamics in CD34+ hematopoietic progenitor cells—separate de novo methylation events and hypomethylation of differentiation-related genes (Bocker et al., 2011). These results show that human stem cells display an aging epigenome phenotype that is analogous to differentiated tissue cells, suggesting a high degree of utility for stem cells in future aging epigenome studies.
Along with research on DNA methylation, there is a need to evaluate environmental deflection in other epigenetic modifications—eg, chromatin state. Recent research has shown that histone marks (ie, H3K4me3, H3k27me3) and chromatin status (heterochromatin vs. euchromatin) are altered with age (Maleszewska et al., 2016; O’Sullivan and Karlseder, 2012). To investigate how age-related changes in chromatin structure interact with DNA methylation and the environment during aging, future studies could investigate longitudinal chromatin state in animal exposure models using repeated Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq) experiments across the life course. Integration of this type of data with longitudinal epigenome-wide DNA methylation and/or histone modification (eg, ChIP-seq) data would allow for a more complete picture of the aging epigenome.
In addition to chromatin state, future aging epigenome studies should also investigate the effects of both aging and the environment on 5-hydroxymethylcytosine (5-hmC). Although traditional bisulfite sequencing approaches do not distinguish 5-hmC from 5-methylcytosine (5-mC), newer technologies—eg, hydroxymethylated DNA immunoprecipitation sequencing (hMeDIP-seq)—are able to specifically measure epigenome-wide 5-hmC levels (Tan et al., 2013). Recent studies have shown that 5-hmC is a stable epigenetic mark enriched at transcription factor binding sites, enhancer regions, and gene regions, but depleted at promoter regions, suggesting a complex role as both a positive and negative regulator of transcription (Stroud et al. 2011; Wu et al., 2011; Hahn et al., 2014; Li et al., 2016). Supporting its role as a regulatory mark, studies have shown that the global loss of 5-hmC is associated with cancer development (Pfeifer et al., 2013), and that 5-hmC is enriched in differentially methylated regions associated with cancer (Li et al., 2016). Active processing of 5-mC to 5-hmC occurs via a Ten-eleven translocation (TET) methylcytosine dioxygenase-mediated oxidative pathway (Shen et al., 2014), and previous studies show that exposure-induced oxidative stress can alter both TET enzyme levels (Coulter et al., 2013) and global hydroxymethylation (Delatte et al., 2015). A recent study in mice showed effects of both alcohol exposure and age on global hydroxymethylation (Tammen et al., 2014), suggesting that DNA hydroxymethylation may also exhibit patterns of environmental deflection. Given that DNA methylation and DNA hydroxymethylation can have distinct directional associations with transcription (Wu et al., 2011), the environmental deflection of either the methylome or hydroxymethylome could have disparate, long-lasting effects on phenotype.
CONCLUSION
In this review, we proposed a new term—environmental deflection—that refers to environment- or toxicant-mediated shifts away from the baseline rates of age-related methylation or epigenetic drift within an organism. By operating through an epigenetic mechanism, environmental factors may impact long-term gene regulation via environmental exposures at sensitive windows of the life course. Thus, environmental deflection of the aging epigenome may underlie the apparent delay between developmental exposure and biological effects later in life, providing further support for the DOHaD hypothesis. A small number of reports, including non-twin human cohort studies and animal model studies, now indicate that exposure to environmental factors can alter the rates of age-related methylation and epigenetic drift across the life course. To further investigate the impact of environmental deflection, we have introduced and described theoretical manifestations of environmental deflection and summarized human and animal studies that support this phenomenon. Toxicant-sensitive epigenetic phenomena that interact with normal aging may help to explain the growing prevalence of chronic diseases in human populations, demonstrating that longitudinal measures of the epigenome should be considered when designing future toxicoepigenetics studies.
FUNDING
This work was supported by the University of Michigan NIEHS/EPA Children's Environmental Health and Disease Prevention Center P01 ES022844/RD83543601, the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center (P30 ES017885), as well as the University of Michigan NIEHS Institutional Training Grant T32 ES007062 (JJK, LM) and the University of Michigan School of Public Health Regents' Fellowship (JJK). The authors have no conflicts of interest and declare no competing financial interests.
REFERENCES
- Alisch R. S., Barwick B. G., Chopra P., Myrick L. K., Satten G. A., Conneely K. N., Warren S. T. (2012). Age-associated DNA methylation in pediatric populations. Genome Res. 22, 623–632. 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson O. S., Sant K. E., Dolinoy D. C. (2012). Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 23, 853–859. 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A., Shamy M. Y., Chervona Y., Clancy H. A., Sun H., Hall M. N., Qu Q., Gamble M. V., Costa M. (2012). The effect of exposure to carcinogenic metals on histone tail modifications and gene expression in human subjects. J. Trace Elem. Med. Biol. 26, 174–178. 10.1016/j.jtemb.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P., Barker D., Clutton-Brock T., Deb D., D'Udine B., Foley R. A., Gluckman P., Godfrey K., Kirkwood T., Lahr M. M., et al. (2004). Developmental plasticity and human health. Nature 430, 419–421. 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- Bell J. T., Tsai P. C., Yang T. P., Pidsley R., Nisbet J., Glass D., Mangino M., Zhai G., Zhang F., Valdes A., et al. (2012). Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 8, e1002629. 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A. J., Jirtle R. L. (2010). Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res. A Clin. Mol. Teratol. 88, 938–944. 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi S. W., Huang H., Wu J., Zawia N. H. (2011). Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer's disease. J Alzheimers Dis. 27, 819–833. 10.3233/JAD-2011-111013 [DOI] [PubMed] [Google Scholar]
- Bocker M. T., Hellwig I., Breiling A., Eckstein V., Ho A. D., Lyko F. (2011). Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117, e182–e189. 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- Boks M. P., van Mierlo H. C., Rutten B. P., Radstake T. R., De Witte L., Geuze E., Horvath S., Schalkwyk L. C., Vinkers C. H., Broen J. C., et al. (2015). Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology 51, 506–512. 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Boumber Y. A., Kondo Y., Chen X., Shen L., Guo Y., Tellez C., Estécio M. R., Ahmed S., Issa J. P. (2008). An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 4, e1000162. 10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody G. H., Yu T., Chen E., Beach S. R., Miller G. E. (2016). Family-centered prevention ameliorates the longitudinal association between risky family processes and epigenetic aging. J. Child Psychol. Psychiatry 57, 566–574. 10.1111/jcpp.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Fernandez J. E., Spector T. D., Bell J. T. (2014). Epigenetics of discordant monozygotic twins: implications for disease. Genome Med. 6, 60. 10.1186/s13073-014-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. H., Marioni R. E., Colicino E., Peters M. J., Ward-Caviness C. K., Tsai P. C., Roetker N. S., Just A. C., Demerath E. W., Guan W., et al. (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 8, 1844–1865. 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N., Wang L., Lu X., Senut M. C., Brenner C., Ruden D. M. (2011). Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6, 853–856. [DOI] [PubMed] [Google Scholar]
- Coulter J. B., O'Driscoll C. M., Bressler J. P. (2013). Hydroquinone increases 5-hydroxymethylcytosine formation through ten eleven translocation 1 (TET1) 5-methylcytosine dioxygenase. J. Biol. Chem. 288, 28792–28800. 10.1074/jbc.M113.491365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K., Waite L. L., Thalacker-Mercer A., West A., Bamman M. M., Brooks J. D., Myers R. M., Absher D. (2013). Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 14, R102. 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B., Jeschke J., Defrance M., Bachman M., Creppe C., Calonne E., Bizet M., Deplus R., Marroquí L., Libin M., et al. (2015). Genome-wide hydroxymethylcytosine pattern changes in response to oxidative stress. Sci. Rep. 5, 12714. 10.1038/srep12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Dzitoyeva S. G., Matrisciano F., Tueting P., Grayson D. R., Guidotti A. (2015). Brain-derived neurotrophic factor epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol. Psychiatry 77, 589–596. 10.1016/j.biopsych.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosunmu R., Alashwal H., Zawia N. H. (2012). Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 133, 435–443. 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P. A. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463. 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Essex M. J., Boyce W. T., Hertzman C., Lam L. L., Armstrong J. M., Neumann S. M., Kobor M. S. (2013). Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Dev. 84, 58–75. 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairweather D. S., Fox M., Margison G. P. (1987). The in vitro lifespan of MRC-5 cells is shortened by 5-azacytidine-induced demethylation. Exp. Cell Res. 168, 153–159. [DOI] [PubMed] [Google Scholar]
- Faulk C., Liu K., Barks A., Goodrich J. M., Dolinoy D. C. (2014). Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9, 934–941. 10.4161/epi.29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M. F., Ballestar E., Paz M. F., Ropero S., Setien F., Ballestar M. L., Heine-Suñer D., Cigudosa J. C., Urioste M., Benitez J., et al. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. U S A 102, 10604–10609. 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert K. M., Blossom S. J., Erickson S. W., Reisfeld B., Zurlinden T. J., Broadfoot B., West K., Bai S., Cooney C. A. (2016). Chronic exposure to water pollutant trichloroethylene increased epigenetic drift in CD4(+) T cells. Epigenomics 8, 633–649. 10.2217/epi-2015-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert K. M., Nelson A. R., Cooney C. A., Reisfeld B., Blossom S. J. (2012). Epigenetic alterations may regulate temporary reversal of CD4(+) T cell activation caused by trichloroethylene exposure. Toxicol. Sci. 127, 169–178. 10.1093/toxsci/kfs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell M. A., Rando T. A. (2015). Stem cells and healthy aging. Science 350, 1199–1204. 10.1126/science.aab3388 [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C., Covert T. R., Haque M. M., Settles M., Nilsson E. E., Anway M. D., Skinner M. K. (2012). Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod. Toxicol. 34, 694–707. 10.1016/j.reprotox.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. A., Szabó P. E., Pfeifer G. P. (2014). 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104, 314–323. 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa N., Oshima M., Sashida G., Matsui H., Koide S., Saraya A., Wang C., Muto T., Takane K., Kaneda A., et al. (2016). Impact of combinatorial dysfunctions of Tet2 and Ezh2 on the epigenome in the pathogenesis of myelodysplastic syndrome. Leukemia [DOI] [PubMed] [Google Scholar]
- Heindel J. J., Balbus J., Birnbaum L., Brune-Drisse M. N., Grandjean P., Gray K., Landrigan P. J., Sly P. D., Suk W., Cory Slechta D., et al. (2015). Developmental Origins of Health and Disease: Integrating Environmental Influences. Endocrinology 156, 3416–3421. 10.1210/EN.2015-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H., Li N., Ferreira H. J., Moran S., Pisano D. G., Gomez A., Diez J., Sanchez-Mut J. V., Setien F., Carmona F. J., et al. (2012). Distinct DNA methylomes of newborns and centenarians. Proc. Natl Acad. Sci. U S A 109, 10522–10527. 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14, R115. 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Erhart W., Brosch M., Ammerpohl O., von Schönfels W., Ahrens M., Heits N., Bell J. T., Tsai P. C., Spector T. D., et al. (2014). Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci. U S A 111, 15538–15543. 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa J. P. (2014). Aging and epigenetic drift: a vicious cycle. J. Clin. Invest. 124, 24–29. 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. J., Goodman S. J., Kobor M. S. (2015). DNA methylation and healthy human aging. Aging Cell 14, 924–932. 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M., Pfeifer G. P. (2015). Aging and DNA methylation. BMC Biol. 13, 7. 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalani A., Kamat P. K., Givvimani S., Brown K., Metreveli N., Tyagi S. C., Tyagi N. (2014). Nutri-epigenetics ameliorates blood-brain barrier damage and neurodegeneration in hyperhomocysteinemia: role of folic acid. J. Mol. Neurosci. 52, 202-15.10.1007/s12031-013-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola N., Ahola A., Maga M., Mallitt K. A., Fahey P., Cox T. C., Whitelaw E., Chong S. (2010). Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 6, e1000811. 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Sartor M. A., Rozek L. S., Faulk C., Anderson O. S., Jones T. R., Nahar M. S., Dolinoy D. C. (2014). Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC Genomics 15, 30. 10.1186/1471-2164-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J., Marchlewicz E. H., Savidge M., Montrose L., Faulk C., Dolinoy D. C. (2016). Longitudinal effects of developmental bisphenol A and variable diet exposures on epigenetic drift in mice. Reprod. Toxicol. 10.1016/j.reprotox.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer B. I., Mantha K., Kleiber M. L., Diehl E. J., Addison S. M., Singh S. M. (2013). Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis. Model. Mech. 6, 977–992. 10.1242/dmm.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque M. L., Casey K. F., Szyf M., Ismaylova E., Ly V., Verner M. P., Suderman M., Brendgen M., Vitaro F., Dionne G., et al. (2014). Genome-wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics 9, 1410–1421. 10.4161/15592294.2014.970060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. E., Lu A. T., Bennett D. A., Horvath S. (2015). Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging (Albany NY) 7, 1198–1211. 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu Y., Salz T., Hansen K. D., Feinberg A. P. (2016). Whole genome analysis of the methylome and the hydroxymethylome in normal and malignant lung and liver. Genome Res. [Epub ahead of print], 10.1101/gr.211854.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J., Baccarelli A., Mittleman M. A., Sparrow D., Vokonas P. S., Tarantini L., Schwartz J. (2012). Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics 7, 63–70. 10.4161/epi.7.1.18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S., Hinkal G., Kim H. S., Shen L., Zhang L., Zhang J., Zhang N., Liang S., Donehower L. A., Issa J. P. (2010). Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20, 332–340. 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszewska M., Mawer J. S. P., Tessarz P. (2016). Histone Modifications in Ageing and Lifespan Regulation. Curr. Mol. Bio. Rep. 2, 26–35. 10.1007/s40610-016-0031-9 [Google Scholar]
- Manikkam M., Tracey R., Guerrero-Bosagna C., Skinner M. K. (2013). Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8, e55387. 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A., Kisliouk T., Tabachnik T., Meiri N., Weller A. (2014). Overweight and CpG methylation of the Pomc promoter in offspring of high-fat-diet-fed dams are not “reprogrammed” by regular chow diet in rats. Faseb J. 28, 4148–4157. 10.1096/fj.14-255620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjonen H., Sierra A., Nyman A., Rogojin V., Gröhn O., Linden A. M., Hautaniemi S., Kaminen-Ahola N. (2015). Early maternal alcohol consumption alters hippocampal DNA methylation, gene expression and volume in a mouse model. PLoS One 10, e0124931. 10.1371/journal.pone.0124931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D., Loke Y. J., Gordon L., Ollikainen M., Cruickshank M. N., Saffery R., Craig J. M. (2013). Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol. 14, R42. 10.1186/gb-2013-14-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva Y. A., Khamis A. M., Kulakovskiy I. V., Ba-Alawi W., Bhuyan M. S., Kawaji H., Lassmann T., Harbers M., Forrest A. R., Bajic V. B., et al. (2014). Effects of cytosine methylation on transcription factor binding sites. BMC Genomics 15, 119. 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt D. M., Knowles B. B., Solter D. (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828. 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh G., Ebrahimi S., Wang S. C., Cortese R., Kaminsky Z. A., Gottesman I. I., Burke J. R., Plassman B. L., Petronis A. (2016). Epigenetic assimilation in the aging human brain. Genome Biol. 17, 76. 10.1186/s13059-016-0946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollikainen M., Smith K. R., Joo E. J., Ng H. K., Andronikos R., Novakovic B., Abdul Aziz N. K., Carlin J. B., Morley R., Saffery R., et al. (2010). DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum. Mol. Genet. 19, 4176–4188. 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- O’Sullivan R. J., Karlseder J. (2012). The great unravelling: chromatin as a modulator of the aging process. Trends in Biochem. Sci. 37, 466–476. 10.1016/j.tibs.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna L., Zhang Y., Mons U., Holleczek B., Saum K. U., Brenner H. (2016). Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics 8, 64. 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Kadam S., Jin S. G. (2013). 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 6, 10. 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard J. F., Puga A. (2010). Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2, 87–104. 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Dean W., Walter J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089–1093. 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Schick S., Fournier D., Thakurela S., Sahu S. K., Garding A., Tiwari V. K. (2015). Dynamics of chromatin accessibility and epigenetic state in response to UV damage. J. Cell Sci. 128, 4380–4394. 10.1242/jcs.173633. [DOI] [PubMed] [Google Scholar]
- Shah S., McRae A. F., Marioni R. E., Harris S. E., Gibson J., Henders A. K., Redmond P., Cox S. R., Pattie A., Corley J., et al. (2014). Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res. 24, 1725–1733. 10.1101/gr.176933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Song C. X., He C., Zhang Y. (2014). Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614. 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S. A., Kelsey G. (2012). De novo DNA methylation: a germ cell perspective. Trends Genet. 28, 33–42. 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Spiers H., Hannon E., Wells S., Williams B., Fernandes C., Mill J. (2016). Age-associated changes in DNA methylation across multiple tissues in an inbred mouse model. Mech. Ageing Dev. 154, 20–23. 10.1016/j.mad.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Feng S., Morey Kinney S., Pradhan S., Jacobsen S. E. (2011). 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 112, R54. 10.1186/gb.2011.12.6.r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K., Takayanagi K., Akimoto A., Ueda K., Shinkai Y., Umezawa M., Takeda K. (2015). Prenatal diesel exhaust exposure disrupts the DNA methylation profile in the brain of mouse offspring. J .Toxicol. Sci. 40, 1–11. 10.2131/jts.40.1. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Yamashita S., Shimazu T., Niwa T., Ushijima T. (2009). The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 19, 1974–1982. 10.1101/gr.093310.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talens R. P., Christensen K., Putter H., Willemsen G., Christiansen L., Kremer D., Suchiman H. E., Slagboom P. E., Boomsma D. I., Heijmans B. T. (2012). Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell 11, 694–703. 10.1111/j.1474-9726.2012.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Xiong L., Xu W., Wu F., Huang N., Xu Y., Kong L., Zheng L., Schwartz L., Shi Y., et al. (2013). Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 41, e84. 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammen S. A., Dolnikowski G. G., Ausman L. M., Liu Z., Sauer J., Friso S., Choi S. W. (2014). Aging and alcohol interact to alter hepatic DNA hydroxymethylation. Alcohol Clin. Exp. Res. 38, 2178–2185. 10.1111/acer.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Plaza M., Tang W. Y., Shang Y., Umans J. G., Francesconi K. A., Goessler W., Ledesma M., Leon M., Laclaustra M., Pollak J., et al. (2014). Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ. Health Perspect. 122, 946–954. 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Menon U., Gentry-Maharaj A., Ramus S. J., Weisenberger D. J., Shen H., Campan M., Noushmehr H., Bell C. G., Maxwell A. P., et al. (2010). Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20, 440–446. 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., West J., Beck S. (2013). Age-associated epigenetic drift: implications, and a case of epigenetic thrift?. Hum. Mol. Genet. 22, R7–R15. 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio R. G., Torró M. I., Bayón G. F., Álvarez-Pitti J., Fernández A. F., Redon P., Fraga M. F., Lurbe E. (2016). Longitudinal study of DNA methylation during the first 5 years of life. J. Transl. Med. 14, 160. 10.1186/s12967-016-0913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J., Nivard M. G., Willemsen G., Hottenga J. J., Helmer Q., Dolan C. V., Ehli E. A., Davies G. E., van Iterson M., Breeze C. E., et al. (2016). Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat. Commun. 7, 11115. 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey K. J., Parnell S. E., Miranda R. C., Golding M. C. (2015). Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenetics Chromatin 8, 39. 10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z., Kimmel J., Totoki K., Hollenbeck E., Reyes T. M. (2010). Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 151, 4756–4764. 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu X., Zhou Y., Xie H., Hong X., Tsai H. J., Wang G., Liu R., Wang X. (2012). Individual variation and longitudinal pattern of genome-wide DNA methylation from birth to the first two years of life. Epigenetics 7, 594–605. 10.4161/epi.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R. A., Jirtle R. L. (2004). Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20, 63–68. [DOI] [PubMed] [Google Scholar]
- Waterland R. A., Michels K. B. (2007). Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 27, 363–388. 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Wells J. C. (2014). Adaptive variability in the duration of critical windows of plasticity: Implications for the programming of obesity. Evol. Med. Public Health 2014, 109–121. 10.1093/emph/eou019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J., Beck S., Wang X., Teschendorff A. E. (2013). An integrative network algorithm identifies age-associated differential methylation interactome hotspots targeting stem-cell differentiation pathways. Sci. Rep. 3, 1630. 10.1038/srep01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V. L., Smith R. A., Ma S., Cutler R. G. (1987). Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 262, 9948–9951. [PubMed] [Google Scholar]
- Wong C. C., Meaburn E. L., Ronald A., Price T. S., Jeffries A. R., Schalkwyk L. C., Plomin R., Mill J. (2014). Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry 19, 495–503. 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. O., Schwartz J., Wright R. J., Bollati V., Tarantini L., Park S. K., Hu H., Sparrow D., Vokonas P., Baccarelli A. (2010). Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. 118, 790–795. 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D'Alessio A. C., Ito S., Wang Z., Cui K., Zhao K., Sun Y. E., Zhang Y. (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684. 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T., Jiao Y., de Jong S., Ophoff R. A., Beck S., Teschendorff A. E. (2015). An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 11, e1004996. 10.1371/journal.pgen.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Xia Y., Bell C. G., Yet I., Ferreira T., Ward K. J., Gao F., Loomis A. K., Hyde C. L., Wu H., et al. (2014). An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat. Commun. 5, 5719. 10.1038/ncomms6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas A. S., Arloth J., Carrillo-Roa T., Iurato S., Röh S., Ressler K. J., Nemeroff C. B., Smith A. K., Bradley B., Heim C., et al. (2015). Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 16, 266. 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]