Abstract
GATA4, a member of the GATA family, serves a key function in several types of cancer, including hepatoblastoma, gastric cancer and breast cancer. However, the function of GATA4 in nasopharyngeal cancer (NPC) is largely unknown. The present study revealed that GATA4 was upregulated in NPC tissue samples and the NPC cell line, 5–8F. Furthermore, the expression of GATA4 was associated with tumor size, metastasis and poor prognosis. Transwell invasion and wound healing analyses demonstrated that GATA4 promoted cell invasion and migration, respectively. Western blotting and reverse transcription-quantitative polymerase chain reaction revealed that GATA4 overexpression decreased the expression of epithelial markers and increased the expression of mesenchymal markers. By contrast, GATA4 inhibition increased the expression of epithelial markers and decreased the mesenchymal markers. Additionally, chromatin immunoprecipitation and dual-luciferase reporter assays revealed that GATA4 promoted epithelial-mesenchymal transition through transcriptionally activating SLUG. Cell counting kit-8 and colony formation assays were performed to analyze the effect of GATA4 on cell proliferation. The results indicated that GATA4 facilitated cell proliferation in NPC. In conclusion, GATA4 acts as an oncogene and serves crucial roles in NPC and GATA4 may find a potential application as therapeutic option in NPC.
Keywords: GATA4, epithelial-mesenchymal transition, metastasis, proliferation, nasopharyngeal cancer
Introduction
Nasopharyngeal cancer (NPC) derives from the epithelial lining of the nasopharynx (1). Although the incidence rate of NPC is relatively low in the majority of the world, representing about 0.7% of the global cancer burden, it is comparatively higher in southern China and Southeast Asia with ~7.3% (2,3). Previous studies have revealed that inheritance, environmental influences and the Epstein-Barr virus serve essential functions in NPC development (4–9).
The GATA family contains GATA1, GATA2, GATA3, GATA4, GATA5 and GATA6 (10). They all contain a zinc finger domain, which can bind to the specific consensus DNA sequence, 5′-A/TGATAA/G-3′ (11). Multiple reports demonstrated that the GATA family serves essential functions in cell proliferation and differentiation (12,13). The GATA family is separated into two subfamilies; the GATA1/2/3 subfamily and the GATA4/5/6 subfamily. Several previous studies have indicated that GATA4 regulates gene transcription by binding to GATA elements (14,15). Additionally, GATA4 was demonstrated to interact with other transcription factors, including Tbx5 and Nkx2-5 (16). GATA4 acts as a transcriptional regulator, regulating hypertrophic growth of the heart (17). Defects or mutations of GATA4 contribute to a series of cardiac problems, including abnormal ventral folding, congenital heart disease as well as hypoplasia of the ventricular myocardium (18). Additionally, GATA4 is crucial to the gastrointestinal, respiratory and reproductive systems and cancer (19). In addition, GATA4 contributes to cancer development and serves as a potent prognostic predictor (20–23). However, the molecular mechanism of GATA4 in NPC remains poorly understood.
Tumor metastasis occurs due to epithelial-mesenchymal transition (EMT) in cancer cells (24,25). EMT is a crucial phenotypic event; it regulates embryonic development, wound healing and tissue remodeling (26). The main characteristics of EMT are decreased epithelial markers and increased mesenchymal markers (26). Several studies have indicated that GATA4 regulates EMT in many types of cancer (27,28). However, the function of GATA4 in NPC remains unknown.
The current study demonstrated that GATA4 was upregulated in NPC and the expression of GATA4 was associated with tumor size, metastasis and pathological grade. Additionally, the upregulation of GATA4 was associated with a poor prognosis in NPC. In addition, GATA4 enhanced EMT through the regulation of SLUG expression in NPC and facilitated cell invasion. Furthermore, cell proliferation was also demonstrated to be regulated by GATA4. In conclusion, the current study indicated that GATA4 acts as an oncogene in NPC.
Materials and methods
Cell culture and tissue samples
The human NPC cell line, 5-8F, was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI 1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin-streptomycin solution at 37°C with 5% CO2. The human immortalized normal nasopharyngeal epithelial cell line, NP69 obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), was cultured in Keratinocyte-SFM medium supplemented with epidermal growth factor (EGF; both Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 5% CO2.
The adjacent normal tissue (ANT; 0.25 cm from tumor tissue) and tumor tissue samples were obtained from patients who were diagnosed with NPC. A total of 42 pairs of ANT and NPC specimens were collected from Department of Otorhinolaryngology, The Second Affiliated Hospital of Zhejiang University School of Medicine (Hangzhou, China) between April 2015 and October 2016. A total of 19 males and 23 females participated in the study with a mean age of 55.3±4.3 years (range, 35–82 years). The current study was approved by The Ethics Committee of The Second Affiliated Hospital of Zhejiang University School of Medicine. All patients provided written informed consent.
Transfection
Cells (3×105) were transfected with 2.5 µg pcDNA3.1 empty vector (Youbio, Hunan, China), 2.5 µg FLAG-GATA4 (pcDNA3.1-GATA4; Youbio) or 30 nM small interfering (si)RNA scrambled (SCR) or GATA4 siRNA (siGATA4; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according the manufacturer's protocol. Following 48 h transfection at 37°C, cells were collected by centrifugation (800 × g, 4°C, 3 min) and used for the further experiments. The sequences of siRNA were as follows: SCR, 5′-UUCUCCGAACGUGUCACGU-3′; siGATA4, 5′-AATCTCGTAGATATGTTTGAC-3′.
Western blotting
Whole protein was extracted from cells and tissues using radioimmunoprecipitation assay buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl and 1% NP-40] with protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). The concentration of protein was measured using a bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.). Subsequently, equal amount of proteins (40 µg) were separated by 8% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. The membranes were blocked by 5% non-fat milk at room temperature for 1 h, and then incubated with primary antibodies at 4°C overnight. The membranes were washed three times and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Finally, blots were visualized using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.). The antibodies used were as follows: Rabbit anti-GATA4 (1:2,000; cat, 19530-1-AP; Proteintech Group, Inc., Chicago, IL, USA), rabbit anti-TWIST (1:1,000; cat: 25465-1-AP; Proteintech Group, Inc.), rabbit anti-a-catenin (1:3,000; cat, 12831-1-AP; Proteintech Group, Inc.), rabbit antibodies from the EMT kit (1:2,000; cat, 9783; Cell Signaling Technology, Inc., Danvers, MA, USA) and mouse anit-β-actin (1:3,000; cat, 6276; Abcam, Cambridge, UK), goat anti-rabbit (HRP-conjugated; 1:5,000; cat, ab205718; Abcam) and goat anti-mouse (HRP-conjugated; 1:3,000; cat, ab205719; Abcam). All the experiments were repeated at least three times.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissue samples and cells using a Qiagen RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. Then, cDNA was reverse transcribed from mRNA at 42°C for 30 min using a PrimeScript RT-PCR kit (Takara Bio, Inc., Otsu, Japan). Subsequently, Power SYBR™ Green PCR Master Mix (Thermo Fisher Scientific, Inc.) was used to perform qPCR analyses in an Applied Biosystems 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling conditions were as follows: 5 min at 98°C, followed by 34 cycles of denaturation at 98°C for 30 sec, annealing at 57°C for 30 sec and extension at 72°C for 35 sec. Primers were as follows: GATA4 forward, 5′-CCCAATCTCGTAGATATGTTTGAC-3′ and reverse, 5′-CCGTTCATCTTGTGGTAGAG-3′; E-cadherin forward, 5′-AAACATCATTGATGCAGACC-3′ and reverse, 5′-GATAGATTCTTGGGTTGGGTC-3′; α-catenin forward, 5′-TGTTACACAGGTTACAACCCT-3′ and reverse, 5′-GCAGCCTTCATCAAATCACC-3′; N-cadherin forward, 5′-CAAAGCCTGGAACATATGTG-3′ and reverse, 5′-GTTTGAAAGGCCATATGTGG-3′; vimentin forward, 5′-CTCCACGAAGAGGAAATCCA-3′ and reverse, 5′-GATTTGTACCATTCTTCTGCCT-3′; Slug forward, 5′-ACACATACAGTGATTATTTCCC-3′ and reverse, 5′-ACTGTAGTCTTTCCTCTTCAT-3′; Snail forward, 5′-TCTAATCCAGAGTTTACCTTCCAG-3′ and reverse, 5′-TGAAGTAGAGGAGAAGGACGA-3′; Twist forward, 5′-GTACATCGACTTCCTCTACC-3′ and reverse, 5′-GAAACAATGACATCTAGGTCTC-3′; ZEB1 forward, 5′-TTACACCTTTGCATACAGAACCC-3′ and reverse, 5′-TTTACGATTACACCCAGACTGC-3′; GAPDH forward, 5′-ATTTCCTGGTATGACAACGA-3′ and reverse, 5′-TTGATGGTACATGACAAGGTG-3′. GAPDH was used as an internal control. Relative mRNA levels were calculated using the 2−ΔΔCq method (29). All the experiments were repeated at least three times.
Transwell invasion assay
A Transwell invasion assay was performed using Transwell chambers (8 µm pore size; Costar; Corning Incorporated, Corning, NY, USA) according to the manufacturer's protocol. In brief, GATA4 was overexpressed or knocked down in 5-8F cells. Following a 48-h transfection, 20,000 cells were placed into the upper chamber, which was coated with Matrigel (BD Biosciences, San Jose, CA, USA). The cells in the upper chamber were maintained with serum-free RPMI medium 1640, the lower plate was filled with RPMI medium 1640 which contained 10% FBS. Following a 16-h incubation at 37°C, cells on the undersurface were fixed with 10% methanol at room temperature for 10 min and stained with 0.5% crystal violet at room temperature for 10 min. Six fields were randomly selected and cells were counted under a light microscope (magnification, ×20). The number of invaded cells was normalized to the control group. All experiments were repeated at least three times.
Dual-luciferase reporter assay
The promoter region sequences of SLUG and TWIST were cloned into a pGL3-basic plasmid (Biofeng, Beijing, China). For the dual-luciferase reporter assay, 5-8F cells were co-transfected with 0.5 µg Renilla, 0, 0.5, 1 or 2 µg pcDNA3.1-GATA4 (Vigene Biosciences, Rockville, MD, USA) and 2 µg pGL3-SLUG or 2 µg pGL3-TWIST using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Renilla was used to normalize the firefly luciferase activity in each well. Following transfection for 24 h at 37°C, the Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA) was used to measure the relative luciferase activity. All the experiments were repeated at least three times.
Cell counting kit-8 (CCK-8) assay
To investigate the effect of GATA4 on cell proliferation in NPC, 5-8F cells transfected with the empty vector, FLAG-GATA4, scramble small interfering (si)RNA (SCR) or GATA4 siRNA (siGATA4) were placed into 96 well plate at a density of 2,000 cells per well and maintained at 37°C. Then, the cell viability rate was measured using CCK-8 (Beyotime Institute of Biotechnology, Haimen, China) at different time points (0, 24, 48 and 72 h). CCK-8 solution (20 µl) was added to each well and incubated at 37°C for 60 min; the absorbance of each well was measured at 450 nm. All the experiments were repeated at least three times.
Colony formation assay
Transfected 5-8F cells (5×103) were seeded in 6-well plates and maintained in RPMI 1640 containing 10% FBS for 2 weeks at 37°C. Cells were then fixed with 10% methanol at room temperature for 10 min and stained with 0.5% crystal violet at room temperature for 10 min. The number of colonies was counted under a light microscope (magnification, ×20). All the experiments were repeated at least three times.
Wound healing assay
A wound-healing assay was performed to determine the effect of GATA4 on cell migration. Briefly, GATA4 was overexpressed or knocked down in 5-8F cells. When the cells were grown to 90–100% confluence in RPMI 1640 at 37°C, a 200-µl pipette tip was used to scratch the cells. Next, cells were incubated with serum-free RPMI 1640 at 37°C and the migration distance was observed under a light microscope (magnification, ×20) at different time points (0 and 36 h). The relative migrating distance was the ratio of migration distance at 36 h and the distance measured at 0 h. All the experiments were repeated at least three times.
ChIP assay
A ChIP assay was performed using an EZ-ChIP kit (Merck KGaA, Darmstadt, Germany) according to the manufacturer's protocol. In brief, 5-8F cells (~3×106) were used at a density of 80–90% confluence and were fixed with 1% formaldehyde (Beyotime Institute of Biotechnology) at 37°C for 30 min. Following the addition of lysis buffer containing RNase A (Merck KGaA), the lysate was sonicated (20 kHz; amplitude, 40%; 20 cycles, 1 sec on and 1 sec off; 4°C) to break cross-linked chromatin into 200 to 1,000-bp fragments, ChIP assays were performed using 5 µl anti-GATA4 antibody (cat. no. 19530-1-AP; Proteintech Group, Inc.) at 4°C overnight. Protein G agarose beads (60 µl) were used to purify antibody-bound DNA fragments at 4°C over 1 h. Beads were then washed with each 1 ml low salt immune complex wash buffer, high salt immune complex wash buffer, LiCl immune complex wash buffer and TE buffer (all Merck KGaA) at 4°C. DNA was isolated from the immunoprecipitates using a DNA kit (Tiangen Biotech Co., Ltd.) and quantified using RT-qPCR analysis with the specific primer pairs: Slug, forward 5′-CTGGATTATGCCTCTGTGAT-3′ and reverse 5′-TGGTATTTATTTGCTGGTAG-3′; twist, forward 5′-AAGGGATGGACCTGAAACGG-3′ and reverse 5′-GGCAAACTGGAAGCAGCAAA-3′. The reaction mixture contained 2 µl sample, 1 µl primer (10 nM), 10 µl 2X MIX buffer (TransGen Biotech Co., Ltd., Beijing, China) and 7 µl RNase-free. RT-qPCR conditions were as follows: 5 min at 98°C, followed by 35 cycles of denaturation at 98°C for 30 sec, annealing at 56°C for 30 sec and extension at 72°C for 20 sec. Input DNA and DNA immunoprecipitated by anti-IgG antibody served as a positive and negative control, respectively. The results were calculated using the 2−ΔΔCq method (29).
Statistical analysis
All data were presented as mean + standard deviation. SPSS (version 18.0; SPSS, Inc., Chicago, IL, USA) was used to analyze data. The association between GATA4 expression and clinical information was analyzed by Pearson's χ2 test. Kaplan-Meier plots and log-rank tests were used to analyze the rates of survival. Comparisons between two groups were analyzed by Student's t-test. Multiple groups were analyzed by one-way analysis of variance followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
GATA4 is upregulated in NPC tissues and the cell line, 5-8F
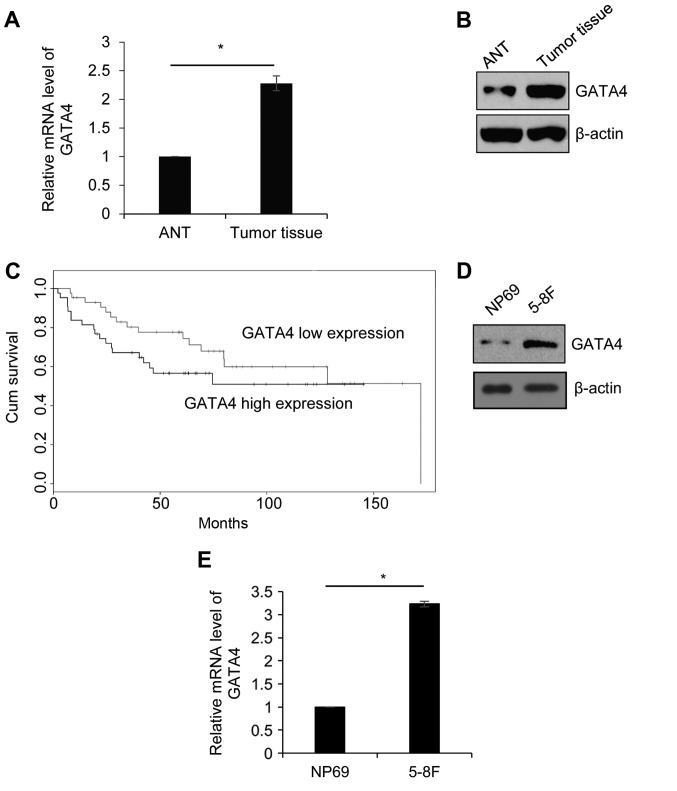
In order to explore the function of GATA4 in NPC, 42 paired NPC tissue and ANT samples were collected from patients who were diagnosed with NPC. The expression of GATA4 was detected by RT-qPCR. As presented in Fig. 1A, the expression of GATA4 in tumor tissue was significantly higher compared with its expression in ANT. Western blotting was performed to detect the protein level of GATA4. Consistent with the RT-qPCR analysis, the expression of GATA4 was markedly higher in tumor tissue compared with ANT (Fig. 1B). Additionally, the association between GATA4 expression and clinicopathology of NPC was analyzed. High GATA4 expression was significantly associated with large tumor size, metastasis and high pathological grade (Table I). In addition, according to survival curves analysis, the patients with high GATA4 expression had lower survival rates compared with the patients who had low GATA4 expression (Fig. 1C). Next, the expression of GATA4 was further investigated in the NPC cell line, 5-8F, and the human immortalized normal nasopharyngeal epithelial cell line, NP69, was used as control. The results demonstrated that the protein and mRNA levels of GATA4 were higher in 5-8F cells compared with that of NP69 cells (Fig. 1D and E). The results indicated that GATA4 serves an essential function in NPC.
Figure 1.
GATA4 is upregulated in nasopharyngeal cancer tissues and the cell line, 5-8F. (A) The relative mRNA expression levels of GATA4 in ANT and tumor tissue samples were detected and quantified using RT-qPCR. (B) GATA4 protein levels in ANT and tumor tissue samples were detected using western blotting (n=42). (C) The association between the expression of GATA4 and the survival rate of patients was analyzed by the Kaplan-Meier method. (D) GATA4 protein levels in NP69 and 5-8F cells were detected using western blotting. (E) The relative mRNA expression levels of GATA4 in NP69 and 5-8F cells were detected and quantified using RT-qPCR. *P<0.05. ANT, adjacent normal tissue; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table I.
Clinicopathological variables in 42 nasopharyngeal cancer patients.
| GATA4 protein expression, n | ||||
|---|---|---|---|---|
| Variables | Patients, n | Low (n=13) | High (n=29) | P-value |
| Sex | 0.555 | |||
| Male | 19 | 5 | 14 | |
| Female | 23 | 8 | 15 | |
| Age, years | 0.588 | |||
| <55 | 20 | 7 | 13 | |
| ≥55 | 22 | 6 | 16 | |
| Tumor size, diameter | 0.038 | |||
| Small (<1 cm) | 16 | 8 | 8 | |
| Large (≥1 cm) | 26 | 5 | 21 | |
| Pathological stage | 0.032 | |||
| I–II | 13 | 7 | 6 | |
| III–IV | 29 | 6 | 23 | |
| Metastasis | 0.015 | |||
| Yes | 30 | 6 | 24 | |
| No | 12 | 7 | 5 | |
Overexpression of GATA4 promotes NPC cell migration and invasion
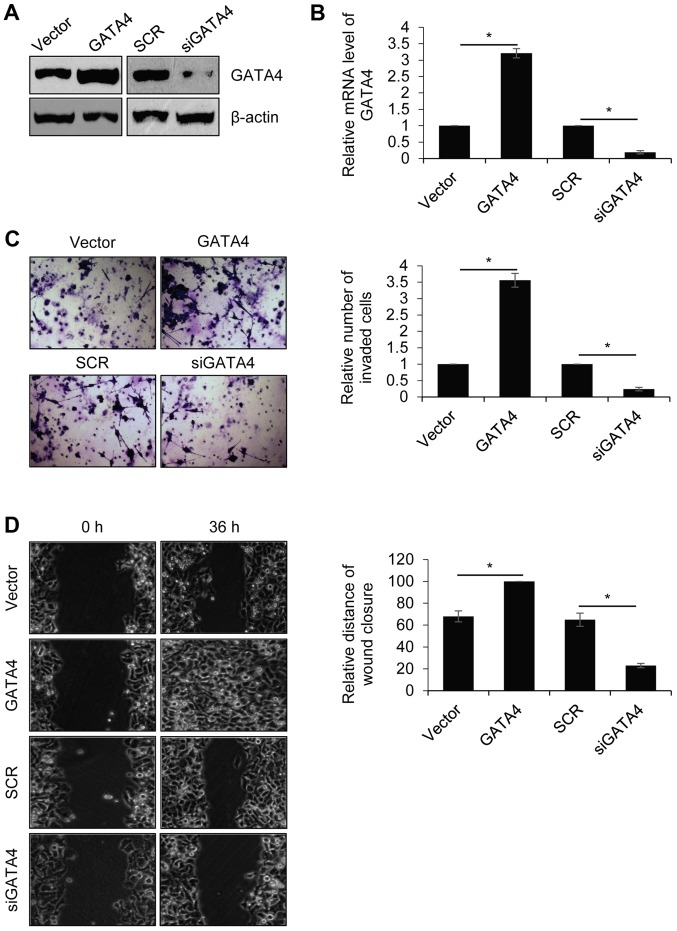
Previous studies by our group indicated that GATA4 was positively correlated with metastasis (22,30). Therefore, the role of GATA4 in the regulation of NPC cell migration and invasion was explored. The human highly metastatic NPC cell line, 5-8F, was used for further experiments. 5-8F cells were transfected with the empty vector, FLAG-GATA4, SCR or siGATA4. Western blotting and RT-qPCR were performed to determine GATA4 expression in 5-8F cells following transfection. The results revealed that the protein and mRNA levels of GATA4 were markedly (Fig. 2A) and significantly (Fig. 2B) increased, respectively, in 5-8F cells transfected with FLAG-GATA4 compared with the vector group. The protein and mRNA levels of GATA4 were markedly and significantly decreased, respectively, in 5-8F cells transfected with siGATA4 compared with the SCR group. Transwell and wound healing assays were performed to determine the effect of GATA4 on cell invasion and migration. As demonstrated in Fig. 2C, overexpression of GATA4 significantly promoted the invasion of 5-8F cells compared with the vector group. Additionally, GATA4 inhibition significantly inhibited the invasion of 5-8F cells compared with the SCR group. In addition, overexpression of GATA4 significantly facilitated cell migration in 5-8F cells compared with the vector group, whereas GATA4 inhibition significantly suppressed cell migration in 5-8F cells compared with the SCR group (Fig. 2D). These results demonstrated that GATA4 promotes NPC cell migration and invasion.
Figure 2.
Overexpression of GATA4 promotes nasopharyngeal cancer cell migration and invasion. 5-8F cells were transfected with an empty vector, FLAG-GATA4, SCR or siGATA4. (A) The protein expression of GATA4 was detected using western blotting. (B) The relative mRNA expression levels of GATA4 were detected and quantified using reverse transcription-quantitative polymerase chain reaction. (C) Transwell invasion and (D) wound healing assays were performed to determine the effect of GATA4 expression on cell invasion and migration, respectively (magnification, ×20). *P<0.05. FLAG-GATA4, vector overexpressing GATA4; si, small interfering RNA; SCR, scramble siRNA; siGATA4, siRNA against GATA4.
GATA4 facilitates EMT in NPC cells
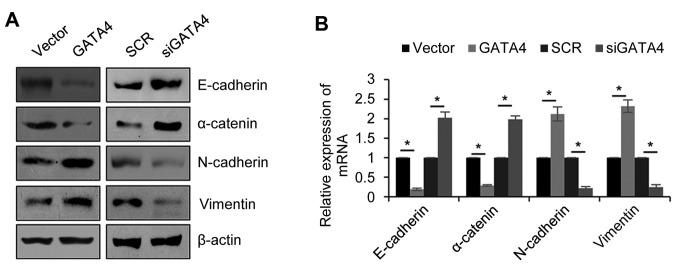
EMT is a complex process, which is associated with metastasis. Therefore, the effect of GATA4 on EMT was explored. The 5-8F cells were transfected with the empty vector, FLAG-GATA4, SCR or siGATA4. Western blotting and RT-qPCR were then used to determine the GATA4 expression in the cells following transfection. As expected, the protein levels of the epithetical markers (E-cadherin and α-catenin) were markedly decreased and mesenchymal markers (N-cadherin and vimentin) were markedly increased when GATA4 was overexpressed compared with the vector group (Fig. 3A). Additionally, knocking down GATA4 expression markedly increased the epithetical markers and decreased the mesenchymal markers compared with the SCR group. The results were similar and significant when the mRNA levels were analyzed (Fig. 3B). These results revealed that GATA4 facilitates EMT in NPC cells.
Figure 3.
GATA4 facilitates epithelial-mesenchymal transition in nasopharyngeal cancer cells. 5-8F cells were transfected with an empty vector, FLAG-GATA4, SCR or siGATA4. (A) Western blotting was used to detect the protein levels and (B) reverse transcription-quantitative polymerase chain reaction analysis was used to detect and quantify the mRNA expression levels of E-cadherin, α-catenin, N-cadherin and vimentin. *P<0.05. FLAG-GATA4, vector overexpressing GATA4; si, small interfering RNA; SCR, scramble siRNA; siGATA4, siRNA against GATA4.
SLUG is transcriptionally activated by GATA4
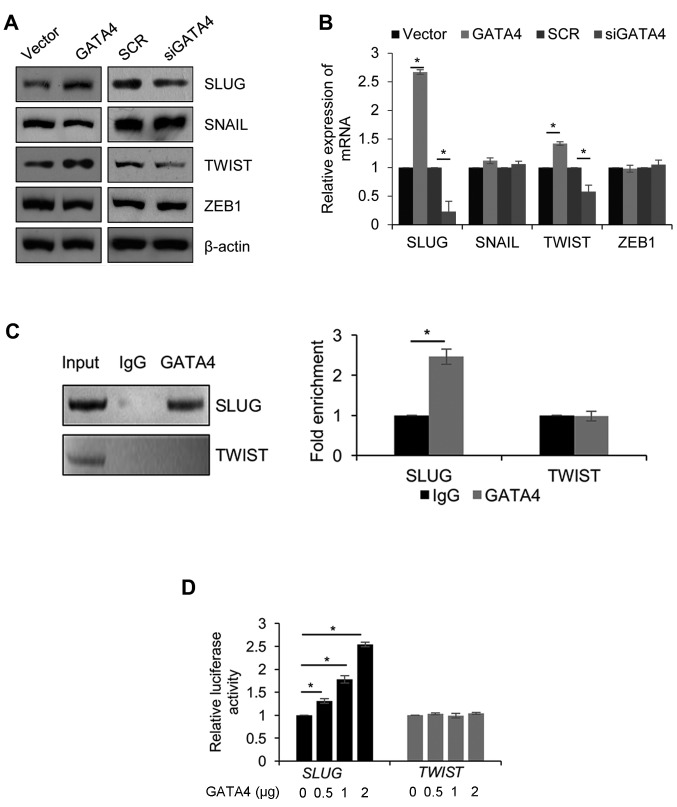
To further decipher the molecular mechanisms of GATA4 in EMT, the expression of genes that serve roles in EMT (SLUG, SNAIL, TWIST and ZEB1) were analyzed to identify whether they are regulated by GATA4. Western blotting demonstrated that the protein expression of SLUG and TWIST was increased when GATA4 was overexpressed; in contrast, the protein expression of SLUG and TWIST was decreased when GATA4 was depleted (Fig. 4A). The results were similar and significant when the mRNA levels were analyzed (Fig. 4B). However, the expression of SNAIL and ZEB1 did not significantly change at the protein and mRNA level. Thus, it was assumed that GATA4 transcriptionally regulated SLUG and TWIST. To validate this hypothesis, a chromatin immunoprecipitation (ChIP) assay was performed to detect the interaction between GATA4, and promoter regions of SLUG and TWIST. The results of the ChIP assay demonstrated that GATA4 directly bound the promoter region of SLUG, but not the promoter region of TWIST (Fig. 4C). Subsequently, a dual-luciferase reporter assay was performed in 5-8F cells, which were co-transfected with pGL3-SLUG or pGL3-TWIST and Renilla, and incubated with increasing concentrations of pcDNA3.1-GATA4. GATA4 significantly increased the luciferase activity of the pGL3-SLUG group, but not in the pGL3-TWIST group (Fig. 4D). These results suggested that GATA4 transcriptionally activates SLUG. Although GATA4 did not transcriptionally regulate TWIST, GATA4 may regulate TWIST through the regulation of other proteins.
Figure 4.
SLUG is transcriptionally activated by GATA4. The (A) protein expression levels and (B) mRNA expression levels of epithelial-mesenchymal transition-associated transcription factors in 5-8F cells transfected with an empty vector, FLAG-GATA4, SCR or siGATA4. Protein expression was detected using western blotting, and mRNA expression was detected and quantified using reverse transcription-quantitative polymerase chain reaction analysis. (C) Chromatin immunoprecipitation analysis was used to detect and quantify the interaction between GATA4 and the promoter regions of SLUG and TWIST in 5-8F cells. (D) A dual-luciferase reporter assay was performed to determine the effect of GATA4 on the transcriptional regulation of SLUG and TWIST. *P<0.05. FLAG-GATA4, vector overexpressing GATA4; si, small interfering RNA; SCR, scramble siRNA; siGATA4, siRNA against GATA4; Ig, immunoglobulin.
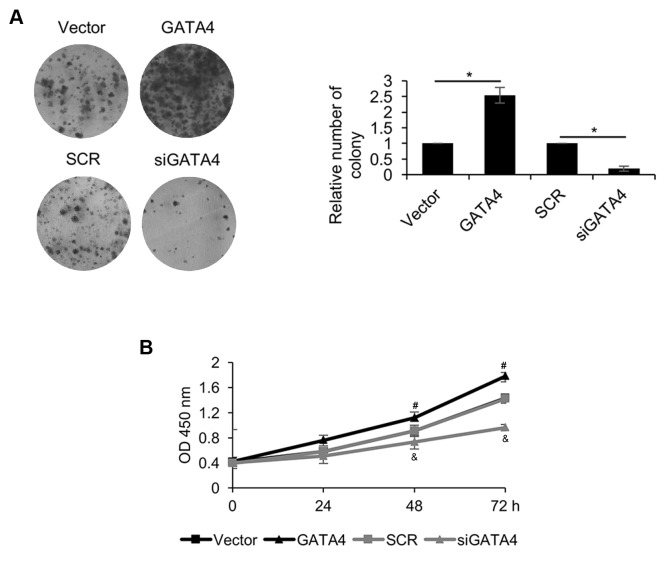
GATA4 promotes cell proliferation in NPC cells
Due to the association between GATA4 expression and tumor size, the role of GATA4 in the regulation of NPC cell proliferation was explored using CCK-8 and colony formation assays. The colony formation assay revealed that overexpression of GATA4 in 5-8F cells significantly increased the number of colonies compared with the vector group, but GATA4 inhibition in 5-8F cells significantly decreased the number of colonies compared with the SCR group (Fig. 5A). The CCK-8 assay demonstrated that overexpression of GATA4 significantly promoted cell proliferation compared with the vector group (Fig. 5B). However, GATA4 inhibition significantly suppressed 5-8F cell proliferation compared with the SCR group. These results suggested that GATA4 promotes cell proliferation in NPC cells.
Figure 5.
GATA4 promotes cell proliferation in NPC cells. (A) Colony formation and (B) Cell counting kit-8 assays quantified cell proliferation of 5-8F cells transfected with an empty vector, a vector overexpressing GATA4, SCR or an siRNA against GATA4 (magnification, ×20). *P<0.05. #P<0.05 vs. the vector group. &P<0.05 vs. the SCR group. si, small interfering RNA; SCR, scramble siRNA; OD, optical density.
Discussion
The results of the current study revealed that GATA4 was upregulated in patients' tissue samples and the 5-8F NPC cell line. In addition, the increased expression of GATA4 in these samples and cells was associated with larger tumor size, high pathological grade, metastasis and poor prognosis. The survival curve analysis indicated that patients with high GATA4 expression survived for ~145 months and patients with low GATA4 expression survived for ~175 months, suggesting that GATA4 may be a novel predictor of NPC. Metastasis is an important event in cancer development, previous reports demonstrated that nasopharyngeal carcinoma frequently led to lymph node metastasis (31,32). As stated above, the current study demonstrated that high GATA4 expression was associated with metastasis. Several functional experiments, including Transwell invasion and wound healing assays, were performed to investigate the mechanism of GATA4 on cell migration and invasion. The results indicated that GATA4 significantly promoted migration and invasion in 5-8F cells.
EMT is a crucial phenotypic event; it regulates embryonic development, wound healing and tissue remodeling (33,34). A number of studies have indicated that GATA4 regulates EMT in several cancers (27,28). However, the role of GATA4 in EMT remains unclear. In order to explore whether GATA4 regulates EMT, GATA4 was overexpressed or knocked down in 5-8F cells. It was demonstrated that the overexpression of GATA4 increased the expression of N-cadherin and vimentin, whereas the expression of E-cadherin and α-catenin was decreased. Conversely, GATA4 inhibition increased the expression of E-cadherin and α-catenin, but the expression of N-cadherin and vimentin was decreased. Therefore, GATA4 promoted EMT in 5-8F cells.
EMT is regulated by several transcription factors, including SLUG, SNAIL, TWIST and ZEB1 (35–38). Subsequently, ChIP and luciferase reporter assays demonstrated that GATA4 transcriptionally activated SLUG. Although previous studies have demonstrated that TWIST is also regulated by GATA4 (39), the current study did not identify GATA4 as a transcriptional regulator of TWIST. Thus, it is assumed that GATA4 may regulate TWIST through other proteins. In summary, GATA4 promotes EMT through transcriptionally activating SLUG in NPC.
Cell proliferation is also an important event in cancer development (40). Due to the association between the expression of GATA4 and tumor size, it was hypothesized that GATA4 may regulate cancer cell proliferation. As expected, the current study revealed that GATA4 facilitated cell proliferation in 5-8F cells.
To the best of our knowledge, the current study is the first study to demonstrate that GATA4 is upregulated in NPC and serves as an oncogene. The high expression of GATA4 predicted a poor prognosis for patients with NPC. Additionally, GATA4 promoted metastasis and proliferation in NPC cells. Therefore, GATA4 may be a novel biomarker for NPC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YZ and BY conceived and designed the present study. YZ, HC and BY performed the experiments. YZ and BY wrote the paper. All authors reviewed and edited the manuscript. The final version of the manuscript was read and approved by all authors, and each author believes that the manuscript represents honest work.
Ethics approval and consent to participate
The current study has been approved by The Ethics Committee of The Second Affiliated Hospital of Zhejiang University School of Medicine. All patients were informed that their tissues would be used in the current study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zeng Z, Huang H, Zhang W, Xiang B, Zhou M, Zhou Y, Ma J, Yi M, Li X, Li X, et al. Nasopharyngeal carcinoma: Advances in genomics and molecular genetics. Sci China Life Sci. 2011;54:966–975. doi: 10.1007/s11427-011-4223-5. [DOI] [PubMed] [Google Scholar]
- 2.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, Hu DX, Tan C, Xiang JJ, Zhou J, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–1974. doi: 10.1158/0008-5472.CAN-03-3253. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Z, Zhou Y, Zhang W, Li X, Xiong W, Liu H, Fan S, Qian J, Wang L, Li Z, et al. Family-based association analysis validates chromosome 3p21 as a putative nasopharyngeal carcinoma susceptibility locus. Genet Med. 2006;8:156–160. doi: 10.1097/01.gim.0000196821.87655.d0. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Z, Huang H, Huang L, Sun M, Yan Q, Song Y, Wei F, Bo H, Gong Z, Zeng Y, et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315–326. doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 7.Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B, Song Y, Li Q, Zeng Y, Liao Q, et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–41782. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q, Li X, Chen P, Xiang B, Zhang W, et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–36799. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Z, Fan S, Zhang X, Li S, Zhou M, Xiong W, Tan M, Zhang W, Li G. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2016;18:206–211. doi: 10.1007/s12094-015-1354-3. [DOI] [PubMed] [Google Scholar]
- 10.Simon MC. Gotta have GATA. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 11.Martin DI, Zon LI, Mutter G, Orkin SH. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 12.Molkentin JD. The zinc finger-containing transcription factors GATA-4, −5, and −6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 13.Ayanbule F, Belaguli NS, Berger DH. GATA factors in gastrointestinal malignancy. World J Surg. 2011;35:1757–1765. doi: 10.1007/s00268-010-0950-1. [DOI] [PubMed] [Google Scholar]
- 14.White RA, Dowler LL, Pasztor LM, Gatson LL, Adkison LR, Angeloni SV, Wilson DB. Assignment of the transcription factor GATA4 gene to human chromosome 8 and mouse chromosome 14: Gata4 is a candidate gene for Ds (disorganization) Genomics. 1995;27:20–26. doi: 10.1006/geno.1995.1003. [DOI] [PubMed] [Google Scholar]
- 15.Engels M, Span PN, Mitchell RT, Heuvel JJTM, Marijnissen-van Zanten MA, van Herwaarden AE, Hulsbergen-van de Kaa CA, Oosterwijk E, Stikkelbroeck NM, Smith LB, et al. GATA transcription factors in testicular adrenal rest tumours. Endocr Connect. 2017;6:866–875. doi: 10.1530/EC-17-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang WY, Heng HH, Liew CC. Assignment of the human GATA4 gene to 8p23.1-->p22 using fluorescence in situ hybridization analysis. Cytogenet Cell Genet. 1996;72:217–218. doi: 10.1159/000134194. [DOI] [PubMed] [Google Scholar]
- 17.Perrino C, Rockman HA. GATA4 and the two sides of gene expression reprogramming. Circ Res. 2006;98:715–716. doi: 10.1161/01.RES.0000217593.07196.af. [DOI] [PubMed] [Google Scholar]
- 18.McCulley DJ, Black BL. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki YJ. Cell signaling pathways for the regulation of GATA4 transcription factor: Implications for cell growth and apoptosis. Cell Signal. 2011;23:1094–1099. doi: 10.1016/j.cellsig.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Färkkilä A, Andersson N, Bützow R, Leminen A, Heikinheimo M, Anttonen M, Unkila-Kallio L. HER2 and GATA4 are new prognostic factors for early-stage ovarian granulosa cell tumor-a long-term follow-up study. Cancer Med. 2014;3:526–536. doi: 10.1002/cam4.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H, Huang S, Zhang X, Wang D, Zhang X, Yuan X, Zhang Q, Huang Z. DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA. Oncol Lett. 2014;8:1751–1756. doi: 10.3892/ol.2014.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi K, Moriguchi T, Miki Y, Nakamura Y, Watanabe M, Ishida T, Yamamoto M, Sasano H, Suzuki T. GATA4 immunolocalization in breast carcinoma as a potent prognostic predictor. Cancer Sci. 2014;105:600–607. doi: 10.1111/cas.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chia NY, Deng N, Das K, Huang D, Hu L, Zhu Y, Lim KH, Lee MH, Wu J, Sam XX, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 24.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 25.Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Tuyn J, Atsma DE, Winter EM, van der Velde-van Dijke I, Pijnappels DA, Bax NA, Knaän-Shanzer S, Gittenberger-de Groot AC, Poelmann RE, van der Laarse A, et al. Epicardial cells of human adults can undergo an epithelial-to-mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem Cells. 2007;25:271–278. doi: 10.1634/stemcells.2006-0366. [DOI] [PubMed] [Google Scholar]
- 28.Campbell K, Whissell G, Franch-Marro X, Batlle E, Casanova J. Specific GATA factors act as conserved inducers of an endodermal-EMT. Dev Cell. 2011;21:1051–1061. doi: 10.1016/j.devcel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Han Q, Xu X, Li J, Wang J, Bai L, Wang A, Wang W, Zhang B. GATA4 is highly expressed in childhood acute lymphoblastic leukemia, promotes cell proliferation and inhibits apoptosis by activating BCL2 and MDM2. Mol Med Rep. 2017;16:6290–6298. doi: 10.3892/mmr.2017.7369. [DOI] [PubMed] [Google Scholar]
- 31.Chan JY, Chow VL, Wong ST, Wei WI. Surgical salvage for recurrent retropharyngeal lymph node metastasis in nasopharyngeal carcinoma. Head Neck. 2013;35:1726–1731. doi: 10.1002/hed.23214. [DOI] [PubMed] [Google Scholar]
- 32.Kang M, Zhou P, Wei T, Zhao T, Long J, Li G, Yan H, Feng G, Liu M, Zhu J, Wang R. A novel N staging system for NPC based on IMRT and RTOG guidelines for lymph node levels: Results of a prospective multicentric clinical study. Oncol Lett. 2018;16:308–316. doi: 10.3892/ol.2018.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Y, Massague J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.e08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci (Landmark Ed) 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- 37.Kojc N, Zidar N, Gale N, Poljak M, Fujs Komlos K, Cardesa A, Höfler H, Becker KF. Transcription factors Snail, Slug, Twist, and SIP1 in spindle cell carcinoma of the head and neck. Virchows Arch. 2009;454:549–555. doi: 10.1007/s00428-009-0771-5. [DOI] [PubMed] [Google Scholar]
- 38.Preca BT, Bajdak K, Mock K, Lehmann W, Sundararajan V, Bronsert P, Matzge-Ogi A, Orian-Rousseau V, Brabletz S, Brabletz T, et al. A novel ZEB1/HAS2 positive feedback loop promotes EMT in breast cancer. Oncotarget. 2017;8:11530–11543. doi: 10.18632/oncotarget.14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoud MM, Kim HR, Xing R, Hsiao S, Mammoto A, Chen J, Serbanovic-Canic J, Feng S, Bowden NP, Maguire R, et al. Twist1 integrates endothelial responses to flow in vascular dysfunction and atherosclerosis. Circ Res. 2016;119:450–462. doi: 10.1161/CIRCRESAHA.116.308870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan B, Ye Y, Liu H, Zhen J, Zhou H, Li Y, Qu L, Wu Y, Zeng C, Zhong W. URG11 regulates prostate cancer cell proliferation, migration, and invasion. Biomed Res Int. 2018;2018:4060728. doi: 10.1155/2018/4060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.