Fig. 3.
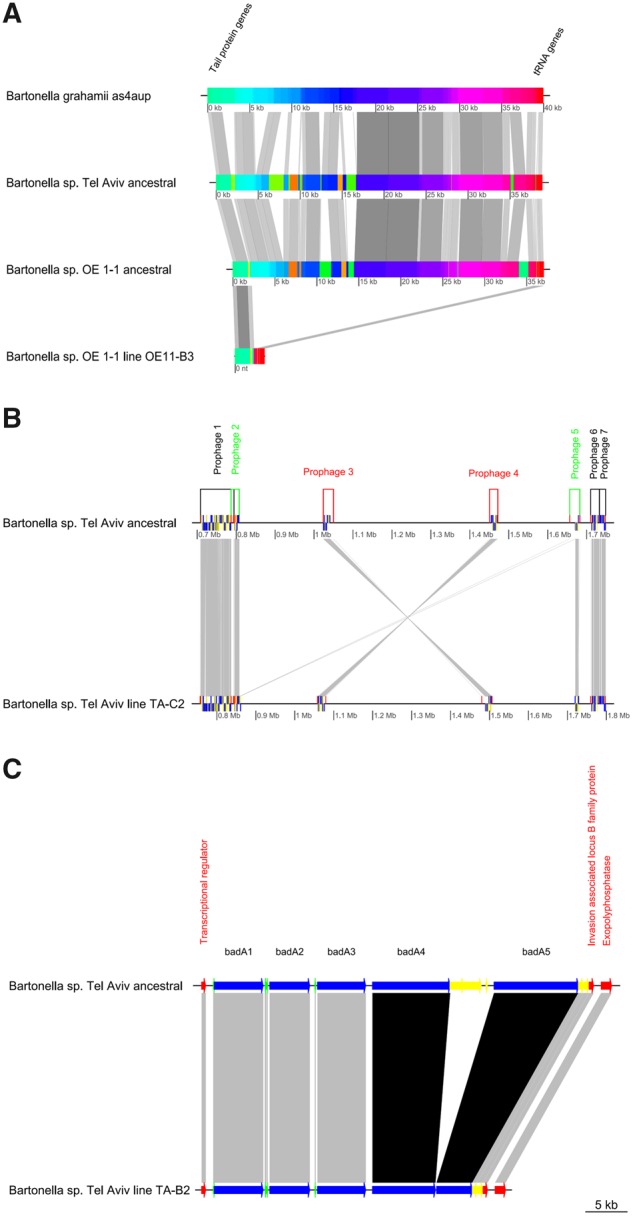
—Structural variation events occurred in prophage and surface protein-gene related regions. (A) MAUVE alignment of homologous prophages from B. grahamii as4aup strain, Bartonella sp. Tel Aviv ancestral genome, Bartonella sp. OE 1-1 ancestral genome (P-BOE11-3) and Bartonella sp. OE 1-1 evolved line OE11-B3 (with deleted prophage). Common genes flaking the loci are indicated in the figure. (B) Comparison of the location of the prophages in Bartonella sp. Tel Aviv ancestral and evolved TA-C2 genomes predicted by PHASTER. Colors indicate: black: prophage in homologous position and orientation; green: prophages that underwent recombination; red: homologous prophages in opposite orientation as a result of an inversion event. (C) Short-read sequencing analysis of Bartonella sp. Tel Aviv ancestor and evolved TA-B2 line (deletion), revealed a ∼13-kb deletion occurred in the evolved line, involving the badA4, badA5 and the intercalated badA remnant genes. All figures were constructed with GenoplotR package in R software.
