Figure 6 |. A model for transcription regulation: precision and plasticity of TRF function achieved via allostery.
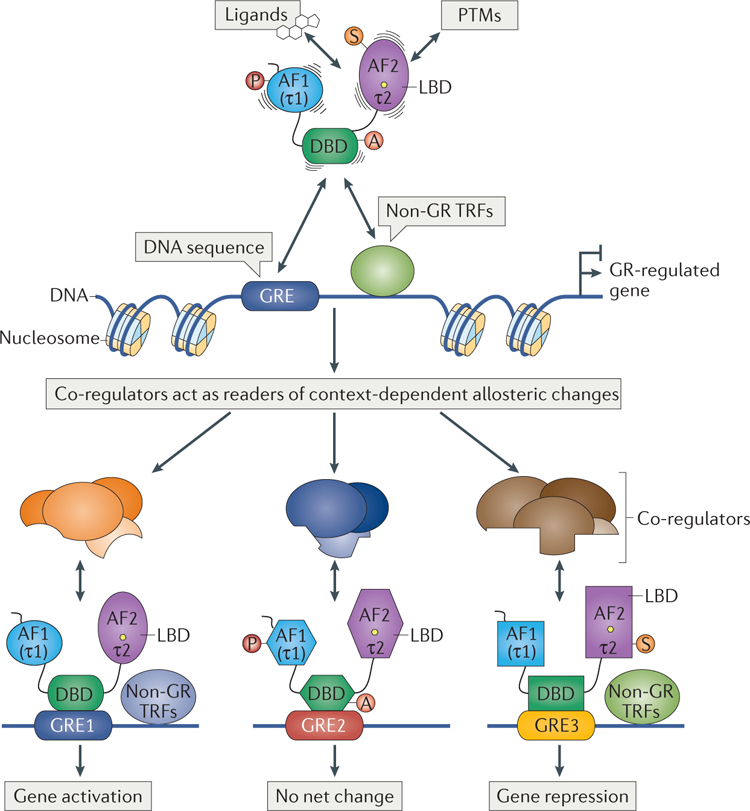
In our model for metazoan transcription regulation, transcriptional regulatory factors (TRFs), such as glucocorticoid receptor (GR), may lack intrinsic regulatory activities, serving instead as molecular scaffolds that can assume different conformations in response to modification by different combinations of specific signalling inputs. Four classes of signalling input are described: ligands and post-translational modifications (PTMs) provide physiological context, DNA-binding seguences provide gene context, and non-GRTRFs provide cell context. Each class of signal confers distinct allosteric effects, and their integrated actions can produce a vast range of GR conformations, which induce, expose or stabilize context-specific proteinsurfacesthat are read by co-regulators. These co-regulators, which are generally large multi-component enzyme-containing complexes, interact in distinct combinations with patterns of cognate GR surfaces and enzymatically modify the general transcriptional machinery and/or surrounding chromatin in and around glucocorticoid response elements (GREs) and target promoters and genes, resulting in the activation or repression of gene expression. Upper panel: lines around GR domains depict direct and allosteric conformational alterations imposed by the signalling inputs. Note that a lack of net regulation (that is, neither activation nor repression) could reflect balanced actions of one or more GREs acting on a single gene (not shown). AF, activation function domain; DBD, DNA-binding domain; LBD, ligand-binding domain. Circles with letters A, P,S indicate different PTMs.
