SUMMARY
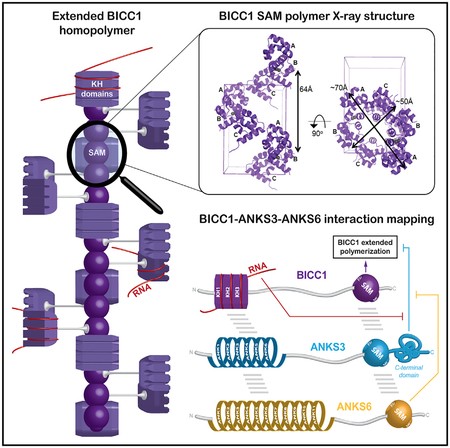
Head-to-tail polymers of sterile alpha motifs (SAM) can scaffold large macromolecular complexes. Several SAM-domain proteins that bind each other are mutated in patients with cystic kidneys or laterality defects, including the Ankyrin (ANK) and SAM domain-containing proteins ANKS6 and ANKS3, and the RNA-binding protein Bicc1. To address how their interactions are regulated, we first determined a high-resolution crystal structure of a Bicc1-SAM polymer, revealing a canonical SAM polymer with a high degree of flexibility in the subunit interface orientations. We further mapped interactions between full-length and distinct domains of Bicc1, ANKS3, and ANKS6. Neither ANKS3 nor ANKS6 alone formed macroscopic homopolymers in vivo. However, ANKS3 recruited ANKS6 to Bicc1, and the three proteins together cooperatively generated giant macromolecular complexes. Thus, the giant assemblies are shaped by SAM domains, their flanking sequences, and SAM-independent protein-protein and protein-mRNA interactions.
Graphical Abstract
In Brief
Rothé et al. describe how ciliopathy-associated proteins Bicc1, ANKS3, and ANKS6 assemble in a mesh-like structure scaffolded by SAM heteropolymers and additional interfaces. These interactions remodel Bicc1 foci size. While ANKS3 disperses them via its C-terminal domain, they can be rescued by recruitment of ANKS6 or a Bicc1 target mRNA.
INTRODUCTION
A growing number of disease-associated mutations are found in genes encoding proteins involved in primary cilia biogenesis and function. These so-called ciliopathies include developmental or later-onset chronic disorders such as nephronophthisis (NPH) and polycystic kidney diseases (PKD) (for review see Hildebrandt et al., 2011). At the apical surface of epithelial cells, primary cilia integrate mechanical, osmotic, and chemical cues to control proliferation, differentiation, and cell polarity through Wnt, cyclic AMP/protein kinase A (cAMP/PKA), Hippo, mechanistic target of rapamycin (mTOR), and other signaling pathways (for review see Walz, 2017). How such diverse stimuli are integrated and transduced by ciliary proteins to control organogenesis and tissue homeostasis is an active area of investigation. Limited space within cilia and specific scaffolds formed by intraflagellar transport proteins may be important to facilitate rapid signal propagation by concentrating specific molecules in close proximity to each other (for review see Nachury, 2014). Signaling molecules may also be locally enriched in specific compartments by “polymerizers” that assemble dynamic platforms called signalosomes (for review see Bienz, 2014). A widespread polymerizer is the sterile alpha motif (SAM), consisting of α-helical bundles of approximately 70 amino acids that can form dimers, closed oligomers, or helical polymers in head-to-tail configuration via so-called mid-loop (ML) and end-helix (EH) surfaces (for review see Qiao and Bowie, 2005). Regularly arrayed macromolecular complexes scaffolded by SAM-domain polymers can accommodate associated proteins or RNAs, but little is known of how the dynamics or signal amplitude of such complexes is regulated (for review see Bienz, 2014).
Mutations in the SAM domains of ANKS6 and Bicc1 are linked to polycystic kidneys (Cogswell et al., 2003; Kraus et al., 2012; Taskiran et al., 2014), whereas ANKS3 mutations have been identified in human patients with autosomal recessive laterality defects, another trait common in several ciliopathies (Shamseldin et al., 2016). ANKS6 contains 11 ANK repeats at its N terminus and a SAM domain close to its C terminus, while ANKS3 consists of 6 N-terminal ANK repeats and a SAM domain followed by 168 amino acids at the C terminus. ANK repeats pack next to each other with a slight torsion angle in rigid platforms to accommodate specific interacting proteins (for review see Mosavi et al., 2004). The functional relevance of ANKS6 in kidney development is highlighted by missense mutations in its SAM domain that cause cystic disease in the PKD cy/+ rat model (Brown et al., 2005), in mouse (Bakey et al., 2015), and in human patients with a nephronophthisis-like disease (Taskiran et al., 2014). In mouse kidneys, ANKS3 immunostaining has been reported in cilia (Delestré et al., 2015), whereas in multi-ciliated Xenopus epidermal cells, GFP-tagged ANKS3 localized at basal bodies and in large cytoplasmic foci (Yakulov et al., 2015). A role in kidney development was demonstrated by ANKS3 knockdown in zebrafish that increased cysts already in the pronephros, the first stage of kidney development (Yakulov et al., 2015).
ANKS3 and ANKS6 interact both with each other and with Bicc1 (Bakey et al., 2015; Delestré et al., 2015; Leettola et al., 2014; Ramachandran et al., 2015; Stagner et al., 2009; Yakulov et al., 2015). A link of Bicc1 to cystic kidneys was discovered in bpk mutant mice, a non-orthologous model of autosomal recessive PKD (ARPKD), where a GC insertion changes the reading frame so that the last 21 amino acids are replaced by 149 aberrant residues (Cogswell et al., 2003). Bicc1 is an RNA-binding protein composed of three KH and two KH-like domains that are linked by an intervening sequence to a C-terminal SAM domain. Targeted deletion of Bicc1 revealed additional roles in the planar positioning of motile node cilia that specify visceral left-right asymmetry and in repressing the translation of adeny-late cyclase 6 (AC6) and PKA inhibitor α(PKIα) mRNAs in kidneys of newborn mice (Maisonneuve et al., 2009; Piazzon et al., 2012). Bicc1 binds the 3′ UTRs of these target mRNAs via KH domains, whereas self-polymerization of its SAM domain mediates recruitment to large cytoplasmic foci for translational silencing (Rothé et al., 2015). Recently, Bicc1 has also been detected in primary cilia of porcine renal epithelial LLC-PK cells (Mohieldin et al., 2015) and at centrosomes of dividing mIMCD3 cells, where it appears to stimulate the translation of specific target mRNAs rather than inhibiting it (Iaconis et al., 2017). Despite 41%–57% sequence identity, the SAM domains of Bicc1, ANKS6, and ANKS3 differ in their potential to self-polymerize. The ANKS6 SAM domain is intrinsically unable to self-interact, whereas recombinant SAM domains of Bicc1 and ANKS3 spontaneously form homopolymers (Knight et al., 2011; Leettola et al., 2014). Self-association via the SAM domain has been confirmed in vivo for full-length Bicc1 (Rothé et al., 2015). A homology-based 3D model of Bicc1-SAM polymers suggests a helical conformation with a pitch of 6 SAM domains (Rothé et al., 2015), in contrast to the crystal structure of the ANKS3-SAM polymer which revealed a pitch of 8 SAM domains per turn (Leettola et al., 2014). How ANKS3 or ANKS6 may affect the conformation or function of Bicc1 homo- or hetero-oligomers is unknown.
Here, we use structural and biochemical approaches combined with yeast two-hybrid (Y2H) assays to comprehensively map interacting domains of Bicc1, ANKS3, and ANKS6, and to assess their potential to hetero-oligomerize and regulate the assembly or activity of cytoplasmic Bicc1. Determination of the crystal structure of homopolymeric Bicc1 SAM domain and measurements of its affinity for recombinant ANKS3 SAM revealed that the ML and EH surfaces of each isolated SAM domain can mediate homo- or hetero-oligomer formation in vitro, consistent with a model of co-polymerization. However, analysis in a cellular context shows that full-length ANKS3 instead disperses large Bicc1 assemblies through its long C-terminal domain, suggesting that this flanking sequence inhibits the elongation of SAM-domain co-polymers. Furthermore, we show that full-length ANKS6 associates with Bicc1 indirectly through ANKS3, thereby rescuing the formation of Bicc1-ANKS3 complexes in large cytoplasmic foci. Our findings suggest that the assembly or disassembly of diverse heteromeric macromolecular SAM-mediated scaffolds is regulated by distinct biophysical properties of both the SAM domains and their relative positioning and specific flanking sequences.
RESULTS
Crystal Structure of the Bicc1 SAM-Domain Polymer
The sequences of Bicc1 and ANKS3 SAM domains are closely related, especially in their ML and EH surfaces (63% and 67% identity, respectively) (Figure 1A). To characterize Bicc1 homo-oligomers, we solved a high-resolution crystal structure using a mutant SAM domain of human Bicc1 where residue R924 was substituted by glutamic acid. This mutation destabilizes the self-association of Bicc1 SAM (see below) enough to solubilize the SAM domain sufficiently for crystallization. Details of the structure determination and refinement are provided in Table 1 and STAR Methods. We solved the structure by multi-wavelength anomalous dispersion (MAD) phasing using a selenomethionine (SeMet)-replaced Bicc1 SAM R924E mutant. The SeMet structure was then used to solve the structure of a native R294E mutant. Since the SeMet structure could be refined to a higher resolution (1.75Å) than the native structure (2.00Å), we employed the SeMet structure for analysis of the SAM-domain interactions. There were three similar but not identical molecules in the asymmetric unit (root-mean-square deviation [RMSD] on all atoms is 0.430–0.631Å) that assemble to form a helical polymer with 6 SAM domains per turn, much like many other SAM domains (Figure 1B). Like other SAM polymers, the subunit interfaces are formed between ML and EH interfaces, but there are unusually large differences in orientation between the non-identical subunits in the crystal, suggesting a high degree of structural flexibility in the Bicc1-SAM polymer (Figures 1B and 1C). The ML surface of Bicc1 consists of negatively charged residues around a shallow hydrophobic patch (Figure 1D, left) that packs against a Phe protrusion surrounded by complementary positively charged residues in the EH surface (Figure 1D, right). Interface residues involved in ionic interactions (D900, D911, K913), hydrogen bonds (E898, K913, K925), hydrophobic packing (F920), and shape complementarity (packing of T903 against the helical backbone of the EH surface) stabilize Bicc1 SAM domain self-interactions (Figure 1D). As electrostatic interactions are adaptable, it is possible that the electrostatically stabilized interface leads to its apparent flexibility. To test whether flexibility may allow SAM polymer architectures to vary, we modeled polymers using each of the three interfaces separately. We found that polymers constructed with only the BA interface have a helical repeat of ~4 SAM domains and a helical pitch of ~48Å. When constructed with only the CB interface, there were ~11 SAM domains per helical repeat with a pitch of 158Å, whereas polymers with only the AC interface harbored ~7 SAM domains per helical repeat with a pitch of 54Å. Thus, it is conceivable that the flexibility to switch interfaces may enable Bicc1 SAM polymers to adjust their conformations to environmental conditions.
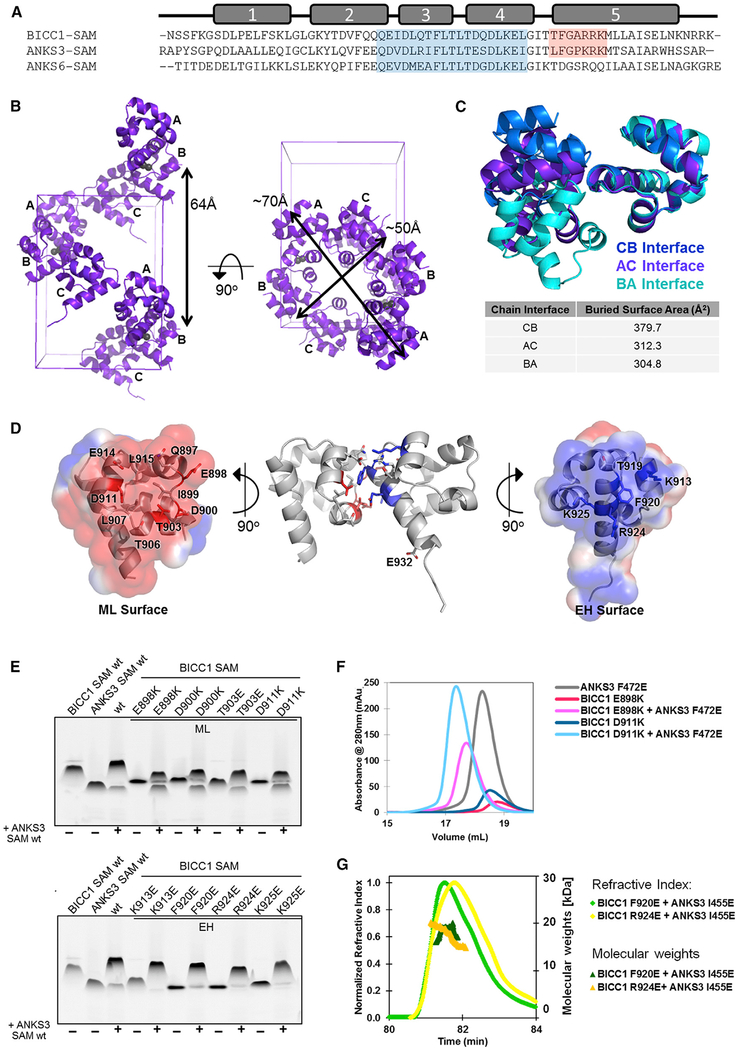
Figure 1. Crystal Structure of the Bicc1 SAM Domain and Binding to ANKS3 SAM.
(A) Sequence alignment of the SAM domains of human Bicc1 (residues 877–938), ANKS3 (residues 429–490), and ANKS6 (residues 771–840). α Helices are numbered above. Residues encompassing ML and EH surfaces are shaded in blue and red, respectively.
(B) Ribbon model of Bicc1 SAM polymer crystal structure. The helical polymer containing 6 subunits per turn is not symmetrical. Each asymmetric unit contains 3 subunits and 2 zinc ions. The zinc ions each have half occupancy and are modeled as gray spheres. A single polymer is shown in the context of the unit cell. The chain ID of each subunit is labeled.
(C) Bicc1 SAMs associate in three different orientations within the polymer. Each homodimer has been aligned using the subunit containing the EH surface, allowing the varying angles of ML surface interaction to be apparent. Surface area buried at each interface was calculated using the PISA server.
(D) Residues critical for Bicc1 SAM interaction are highlighted on the AC interface. Surface electrostatics calculated using APBS in PyMOL and contoured at ±1 kT/e show the charge complementarity between the negatively charged ML surface and positively charged EH surface. The dimer structure (middle) contains the R924E mutation. In the EH surface residue 924 is modeled as the wild-type Arg to more clearly show the positive charge of this surface.
(E) negGFP native gel analysis of Bicc1 SAM and ANKS3 SAM alone (first two lanes) or together (third lane). Residues colored red or blue in (D) were validated as being crucial for the ML or EH surfaces, respectively.
(F) Analytical SEC analysis of the Bicc1-EH/ANKS3-ML interface. Compared with monomeric ML-mutant Bicc1 E898K and D911K and monomeric EH-mutant ANKS3 F472E alone, 1:1 mixtures of the two proteins elute earlier, consistent with heterodimer formation.
(G) SEC-MALS analysis of complexes of ML-mutant ANKS3 I455E with EH-mutant Bicc1 F920E or R924E, respectively, revealed molecular weights of 17.4 ± 0.6 or 18.4 ± 0.1 kDa, corresponding in size to heterodimers (expected molecular weight 16.1 kDa) formed by the wild-type Bicc1-ML/ANKS3-EH interface.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| BICC1 R924E SeMet | BICC1 R924E Native | |||
|---|---|---|---|---|
| PDB ID | 4RQM | 4RQN | ||
| Data collection location | APS 24-ID-C | APS 24-ID-C | ||
| Space group | P212121 | P212121 | ||
| peak | high remote | inflection | ||
| Wavelength (Å) | 0.9791 | 0.9714 | 0.9793 | 0.9795 |
| Cell dimensions | ||||
| a, b, c (Å) | 46.81, 64.43, 70.64 | 46.26, 64.03, 70.05 | 46.25, 64.04, 70.07 | 41.68, 59.22, 68.66 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 1.75 | 2.40 | 2.40 | 2.00 |
| Rsym | 0.068 (0.550) | 0.094 (0.784) | 0.096 (0.866) | 0.097 (0.505) |
| I/σI | 10.29 (2.05) | 8.13 (1.93) | 7.78 (1.74) | 14.94 (4.38) |
| CC1/2 | 99.5 (67.7) | 99.3 (77.1) | 99.4 (75.0) | 99.6 (95.2) |
| Completeness (%) | 93.0 (92.3) | 97.2 (94.6) | 97.3 (93.7) | 98.5 (98.7) |
| Redundancy | 2.27 (2.15) | 2.78 (2.76) | 2.77 (2.65) | 12.65 (13.40) |
| Phasing Statistics | ||||
| No. of sites | 3 | |||
| Mean figure of merit, MAD/after density modification | 0.602/0.778 | |||
| MapCC (SHELXE) | 0.822 | |||
| CC (%) | 65.46 | |||
| Refinement | ||||
| Resolution (Å) | 1.75 | 2.00 | ||
| No. of reflections | 22,189 | 11,813 | ||
| Rwork/Rfree | 0.2297/0.2672 | 0.2124/0.2652 | ||
| No. of atoms | ||||
| Protein | 1,461 | 1,397 | ||
| Water | 40 | 16 | ||
| Zinc | 2 | 1 | ||
| B factor (Å2) | 43.0 | 54.1 | ||
| RMSDs | ||||
| Bond lengths (Å) | 0.008 | 0.008 | ||
| Bond angles (°) | 1.12 | 1.08 | ||
Highest-resolution shell is shown in parentheses. Rsym = Σ|I − <I>|/Σ<I> where I is the observed intensity and <I> is the average intensity from observations of symmetry-related reflections. CC1/2 = correlation coefficient between two halves of the data (Karplus and Diederichs, 2012).
Rwork = Σ|Fobs − Fcalc|/ΣFobs where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated for a set of reflections (10%) that were not included in atomic refinement.
To validate that the structure seen in the crystal corresponds to the polymer in solution, we performed mutational analysis using a native gel assay (Knight et al., 2011). In this assay we fused the SAM domain to a negatively charged GFP (negGFP), which provides consistent migration toward the cathode. Polymeric SAM-negGFP fusions migrate more slowly on a native gel than monomers. As shown in Figure 1E, ML surface mutations E898K, D900K, T903E, and D911K, and the EH surface mutations K913E, F920E, R924E, and K925E each increased the mobility of negGFP fusion proteins on native gels, indicating that they destabilized Bicc1 homopolymers as expected from the crystal structure, even though not all the residues make contacts in every orientation in the polymer.
Bicc1 and ANKS3 SAM Domains Associate in a Manner Compatible with Co-polymerization
The structure of the Bicc1 SAM domain strongly resembles that of ANKS3, showing an average RMSD across backbone atoms of 0.754Å, and of 0.814Å across all atoms (Leettola et al., 2014). Since Bicc1 and ANKS3 have recently been shown to interact (Ramachandran et al., 2015; Stagner et al., 2009; Yakulov et al., 2015), we tested whether negGFP fusions of their SAM domains bind each other in gel-shift assays. We found that negGFP-tagged Bicc1 SAM and negGFP-tagged ANKS3 SAM exhibited diminished electrophoretic mobility upon 1:1 mixing of the two proteins, indicating robust binding (Figure 1E). To identify the protein surfaces mediating this interaction, we mixed ANKS3 SAM 1:1 with Bicc1 SAM domains containing specific point mutations in either the ML or EH surface. Irrespective of whether the Bicc1 SAM domain was mutated in its ML or EH surface, it still shifted ANKS3 (Figure 1E), suggesting that either of these surfaces alone are sufficient to bind the SAM domain of ANKS3. In other words, ANKS3-SAM can bind to both surfaces. To validate this conclusion, we mixed the Bicc1 ML mutants E898K and D911K 1:1 with ANKS3 EH mutant F472E for analysis by size-exclusion chromatography (SEC). A definite elution upshift confirmed heterodimer formation (Figure 1F). We also tested functionality of the alternative interface by mixing the Bicc1 EH mutants F920E and R924E with the ANKS3 ML mutant I455E. Analysis of the resulting complexes by SEC combined with multi-angle light scattering (SEC-MALS) revealed in each case a single peak, eluting at molecular weights of 17.4 ± 0.6 kDa (Bicc1 R924E/ANKS3 I455E) or 18.4 ± 0.1 kDa (Bicc1 F920E/ANKS3 I455E), respectively, consistent with the predicted size of heterodimers (16.1 kDa) (Figure 1G). These results show that the Bicc1 and ANKS3 SAM domains can bind each other via two distinct interfaces: Bicc1-EH + ANKS3-ML and Bicc1-ML + ANKS3-EH.
To measure the strength of these interactions by SPR, we first immobilized the Bicc1 EH mutant R924E and added increasing concentrations of the ANKS3 ML mutant I455E in the mobile phase, allowing only their non-mutated native surfaces to interact. The binding affinity of this Bicc1-ML/ANKS3-EH interface was 2.2 ± 0.1 μM (Figure S1A). Conversely, SPR analysis of immobilized ANKS3 EH mutant F472E with increasing concentrations of the Bicc1 ML mutants E898K or D911K revealed a binding affinity of 1.35 ± 0.1 μM for the Bicc1-EH/ANKS3-ML interface (Figures S1B and S1C). This interaction was specific, since ANKS3 EH mutant F472E did not bind Bicc1 E898K/F920E or D911K/F920E where both ML and EH surfaces are disrupted (Figure S1D). Since the affinities of both possible interfaces are on the same order of magnitude, and are similar to the measured affinity of Bicc1 SAM for itself (1.35 ± 0.1 μM) (Figures S1E–S1G), the Bicc1 and ANKS3 SAM domains may form alternating co-polymers. Alternatively, the EH surface of ANKS3 can strongly bind the ML surface of ANKS6 (KD = 249 ±8 nM) (Leettola et al., 2014). Given its similarity, the EH surface of Bicc1 should bind ANKS6 as well. We tested this hypothesis using negGFP fusion proteins. When the wild-type SAM domains of Bicc1 and ANKS6 were mixed 1:1 there was no gel shift, indicating that they failed to interact under the conditions examined (Figure S1H). Therefore, ANKS6 seems less likely than ANKS3 to form SAM-domain co-polymers with Bicc1.
Semi-quantitative Analysis of Intramolecular Self-Interactions by Yeast Two-Hybrid Assays
To map intra- and intermolecular interactions of Bicc1, ANKS3, ANKS6, and their respective domains, we conducted Y2H assays in Saccharomyces cerevisiae where neither Bicc1 nor ANKS3 or ANKS6 are conserved (Figures 2A–2I and S2). In yeast, interactions between a bait (fused to the DNA-binding domain of Gal4, Gal4-BD) and a prey (fused to the Gal4-activation domain, Gal4-AD) is revealed by selective growth conferred by induction of the HIS3 reporter gene, while titration of 3-amino-1,2,4-triazole as a competitive inhibitor of the HIS3 gene product serves to estimate the strength of each interaction. Each full-length protein (-FL) and individual domains were tested as bait and prey to reduce false-negative results. Empty prey plasmid was used to detect false positives and the threshold below which interactions were considered non-specific. Y2H assays unexpectedly revealed significant binding of Bicc1-FL to its own KH domains (Figures 2A and 2B), and confirmed that the Bicc1 SAM domain strongly interacts with itself (Figure 2C). By contrast, self-interactions of ANKS3-SAM or ANKS6-SAM were weak or undetectable, respectively (Figures 2F and 2I). These results are consistent with prior results indicating that the SAM domain of Bicc1 is more prone to self-polymerize than that of ANKS3, and that the ANKS6 SAM is monomeric (Knight et al., 2011; Leettola et al., 2014).
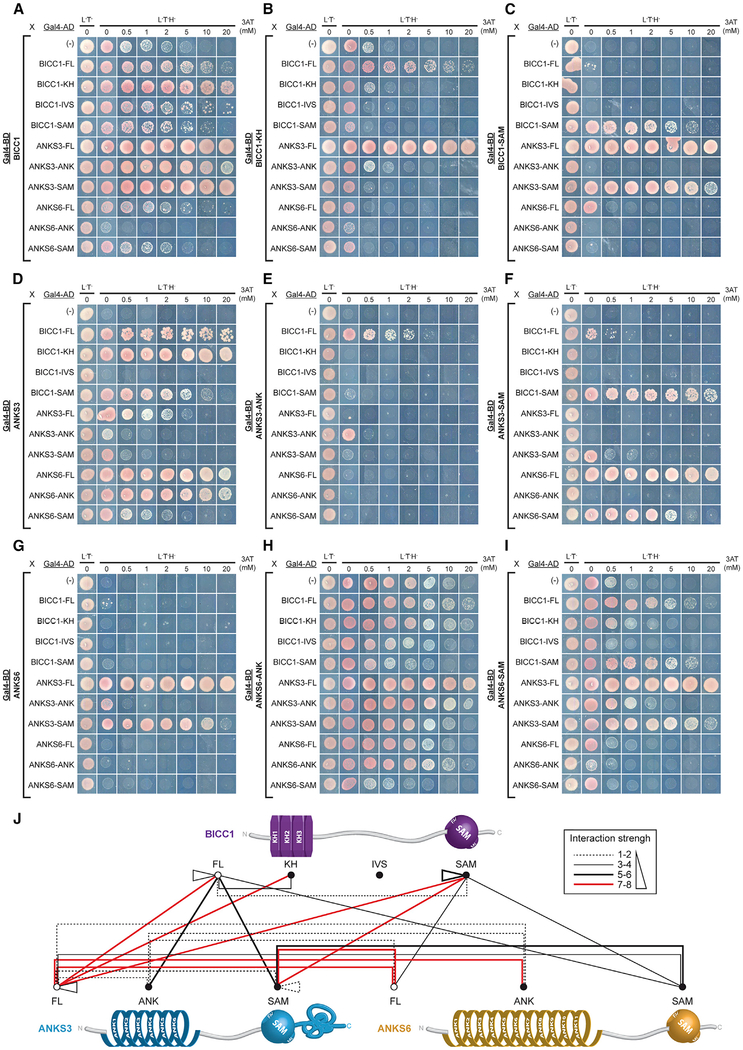
Figure 2. Y2H Mapping of the Bicc1-ANKS3-ANKS6 Interaction Network.
Full-length (FL) human Bicc1, truncated KH (residues 1–414), IVS (415–872), and SAM-domain (873–974) fragments, and full-length ANKS3 and ANKS6 and their respective ANK (34–220 and 8–423, respectively) and SAM (425–488 and 773–836, respectively) domains were used as baits and preys in yeast two-hybrid (Y2H) assays. A growth control in non-selective medium without leucine (L) and tryptophan (T) is shown in the first column (L‒T‒). Interactions are revealed in triple selective medium (L‒T‒H‒) lacking histidine, supplemented with the indicated increasing amounts of 3-aminotriazole (3-AT).
(A–I) Each panel shows one bait protein fused to the DNA-binding domain of Gal4 (Gal4-BD) and the strength of its interactions with all candidate preys fused to Gal4 activation domain (Gal4-AD).
(J) Summary of protein interactions observed in Y2H assays. 3-AT resistance values determined for each interaction were used to define an interaction strength scale (box) from the weakest (thin dotted black line) to the strongest (thick solid red line).
Y2H Reveals an Intricate Interaction Network between Bicc1, ANKS3, and ANKS6
Y2H analysis of Bicc1, ANKS3, and ANKS6 in pairs revealed that full-length Bicc1 and ANKS3 and their respective SAM domains bind each other (Figures 2A, 2C, 2D, and 2F). Strong interactions were also detected between ANKS3 and ANKS6 and between their SAM domains (Figures 2D, 2F, 2G, and 2I). In sharp contrast, only minimal binding was detected between isolated SAM domains of Bicc1 and ANKS6 (Figures 2C and 2I) or the corresponding full-length proteins (Figures 2A and 2G). These data confirm the conclusion of our gel-shift assay and extend it to full-length proteins, namely that the Bicc1 SAM domain preferably binds the SAM domain of ANKS3 and not ANKS6. Additional robust interactions between Bicc1 and ANKS3 involved their ANK and KH domains. Especially the ANK repeats of ANKS3 strongly bound Bicc1-FL both as bait and as prey (Figures 2A and 2E) and, conversely, the KH domain of Bicc1 bound ANKS3-FL (Figures 2B and 2D). However, Bicc1-KH and ANKS3-ANK domains alone failed to directly interact (Figures 2B and 2E), suggesting that additional determinants are required for their stable association. Equally unexpected, ANKS3-FL strongly interacted with ANKS6-ANK (Figure 2D). This interaction was confirmed using ANKS6-ANK as bait despite increased background and, conversely, by significant binding of ANKS6-FL as bait with ANKS3-ANK (Figure 2G). Together, these observations suggest that Bicc1-ANKS3 and ANKS3-ANKS6 complexes rely both on SAM-SAM interaction, as well as additional binding surfaces involving ANK repeats and KH domains (Figure 2J).
ANKS3 Recruits ANKS6 to Bicc1
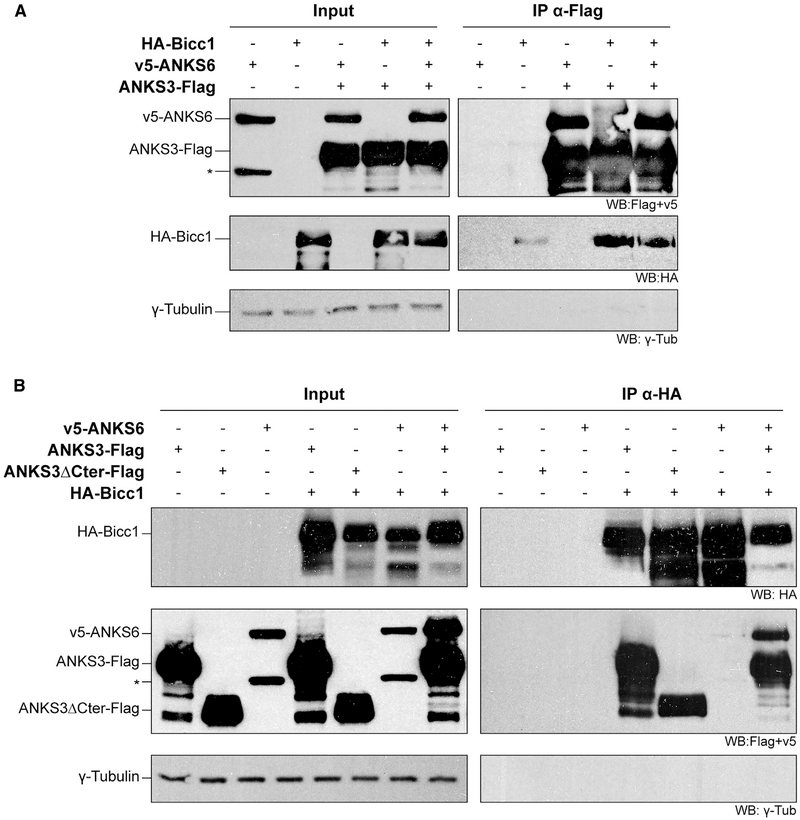
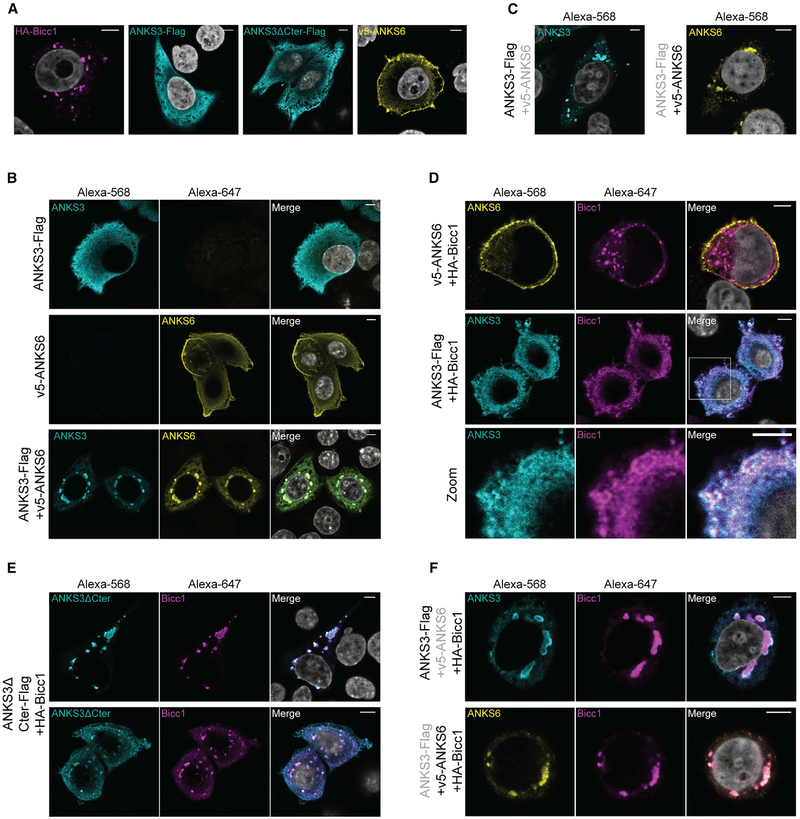
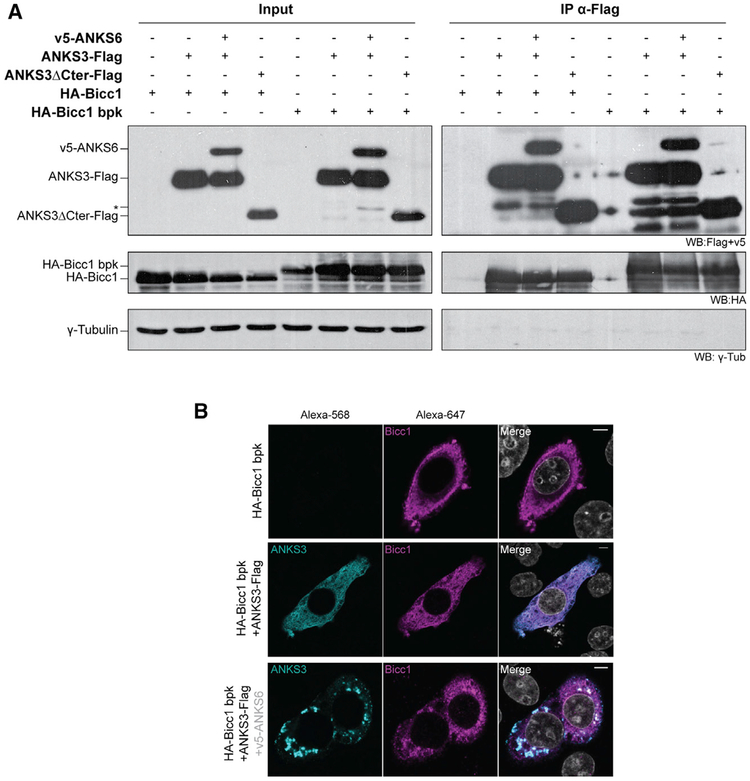
Since ANKS6 associated with ANKS3 but not with Bicc1 in Y2H assays, it may be recruited to Bicc1 by ANKS3. To test this hypothesis, we used HEK293T cells that present the advantage of not expressing endogenous Bicc1, and performed co-immunoprecipitation assays by co-expressing HA-Bicc1 or v5-ANKS6, or both, with ANKS3-Flag. Irrespective of whether HA-Bicc1 and v5-ANKS6 were co-expressed jointly or individually, they each co-immunoprecipitated with ANKS3-Flag (Figure 3A). Thus, Bicc1 and ANKS6 do not appear to compete with each other for binding to ANKS3. Conversely, an HABicc1 pull-down efficiently enriched ANKS3-Flag as well as an ANKS3-Flag truncation mutant lacking the C-terminal 168 amino acids (ANKS3ΔCter-Flag) (Figure 3B). These results confirm Bicc1-ANKS3 binding and show that it is independent of the ANKS3 C-terminal domain. In sharp contrast to ANKS3-Flag, v5-ANKS6 barely co-immunoprecipitated with HABicc1, in line with two-hybrid assays showing that ANKS6 and Bicc1 only weakly interact. Possibly, small amounts of endogenous ANKS3 stabilize this interaction, because the presence of ANKS3-Flag greatly enriched v5-ANKS6 in HA-Bicc1 immunoprecipitates.
Figure 3. Bicc1-ANKS6 Association Is Stabilized by ANKS3.
(A) HA-Bicc1 and v5-ANKS6 co-immunoprecipitation with ANKS3-FLAG in HEK293T cell extracts by anti-FLAG beads. Asterisk denotes a second ANKS6-specific band of reduced size.
(B) ANKS3-FLAG, ANKS3ΔCter-FLAG, and v5-ANKS6 co-immunoprecipitation with HA-Bicc1 by anti-HA beads. Input was 5% of total cell extracts. γ-Tubulin was used as negative control. Asterisk as in (A).
Analysis of Bicc1 Homo- and Hetero-oligomers by Sucrose Density Gradient Fractionation Confirms ANKS3-Mediated ANKS6 Recruitment in High-Molecular-Weight Complexes
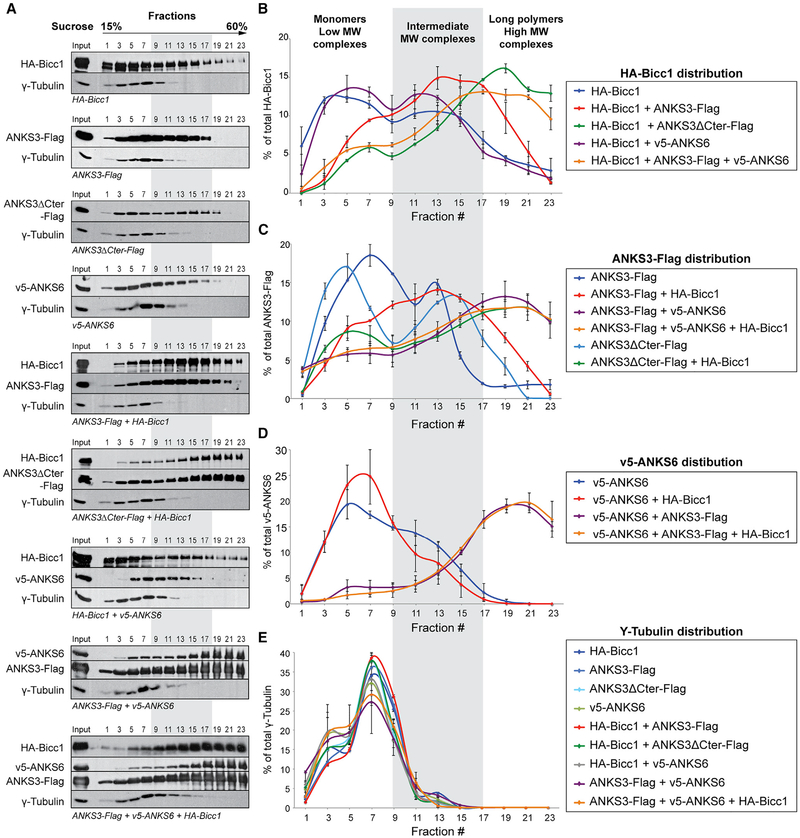
To confirm whether ANKS6 recruitment to Bicc1 is mediated by ANKS3, we analyzed oligomeric complexes by sucrose density gradient fractionation and their repartitioning to different density fractions after co-transfection in HEK293T cells. This approach allows to discriminate complexes by their size and to evaluate how each subunit influences the protein network extension. Co-transfection of Bicc1 with ANKS3 shifted their respective curves from two peaks to a single one with a maximum in the intermediate molecular weight range around fraction 13 (Figures 4A–4C). Interestingly, deletion of the C-terminal domain of ANKS3 shifted ANKS3 and Bicc1 complexes even further to the high molecular weight (HMW) range and in amounts that far exceeded those observed without exogenous ANKS3 (43% versus 11%). The same HMW fractions 19–23 also enriched ANKS3 and ANKS6 if both proteins were co-expressed with or without Bicc1, but not if either of them was transfected alone (Figures 4A–4D). In sharp contrast, no interference was observed between Bicc1 and ANKS6 (Figures 4A, 4B, and 4D), arguing that the effects of ANKS3 were highly specific. As an additional control, the distribution of γ-tubulin remained unchanged in all conditions examined, confirming correct sucrose fractionation (Figure 4E). Thus, Bicc1, ANKS6, and ANKS3 form complexes of dramatically diverse sizes, depending on their combination and on the C-terminal domain of ANKS3, which inhibits the recruitment of Bicc1 into hetero-oligomeric HMW complexes.
Figure 4. Density Gradient Fractionation Reveals Complexes of Diverse Sizes and Composition.
(A) Combinations of HA-Bicc1, ANKS3-Flag, ANKS3ΔCter-Flag and v5-ANKS6 were transiently expressed in HEK293T cells. Cell extracts were fractionated on a continuous15%–60% sucrose gradient and analyzed by western blotting. The migration from the top to the bottom is indicated. γ-Tubulin was used as internal control.
(B–E) Graphs showing the percentage of protein for each fraction compared with the cumulated signal in the whole gradient. Error bars show SEM.
Full-Length ANKS3 via Its C-Terminal Domain Can Inhibit Cytoplasmic Clustering of Bicc1
Since HA-Bicc1 and ANKS3-Flag co-fractionated in sucrose gradient, and because recruitment of ANKS6 increased the apparent average size of HA-Bicc1 and ANKS3-Flag in HMW complexes, we asked whether these effects correlate with increased Bicc1 clustering in cytoplasmic foci. To address this question, we analyzed combinations of HA-Bicc1, ANKS3-Flag and v5-ANKS6 in HeLa cells by indirect immune-fluorescent staining. As shown previously, HA-Bicc1 formed large cytoplasmic clusters, reflecting its strong capacity to polymerize (Maisonneuve et al., 2009; Rothé et al., 2015). By contrast, ANKS3-Flag alone localized diffusely throughout the cytoplasm, and v5-ANKS6 alone was enriched at cell protrusions and at the cortex beneath the plasma membrane (Figure 5A). To evaluate effects of full-length proteins on each other’s localization, we co-expressed ANKS3-Flag with v5-ANKS6, followed by immunostaining with mouse anti-Flag and rabbit anti-ANKS6 antibodies in combination (Figure 5B) or with mouse anti-Flag or mouse anti-v5 individually (Figure 5C). ANKS3 and ANKS6 drastically changed their distinct individual distributions and co-localized in large cytoplasmic foci only if they were co-expressed. ANKS3-Flag also translocated to cytoplasmic foci when co-expressed with HA-Bicc1, although these were extremely small compared with foci of Bicc1 alone or of ANKS3-ANKS6 complexes (Figure 5D). To assess how ANKS3 disperses large Bicc1 foci, we compared it with mutant ANKS3ΔCter-Flag. Contrary to wild-type ANKS3, truncated ANKS3ΔCter-Flag co-localized with HABicc1 exclusively in large or medium-size cytoplasmic foci in 90% and 10% of the cells, respectively (n = 60, Figure 5E). Only the smaller of these percentiles showed significant diffuse staining of both proteins also outside foci throughout the cytoplasm (Figure 5E, bottom panel). This result shows that ANKS3 disperses Bicc1 foci specifically through its C-terminal domain. In sharp contrast to ANKS3-Flag, v5-ANKS6 did not translocate to HA-Bicc1 foci (Figure 5D). This result confirms our earlier conclusion that ANKS6 and Bicc1 do not directly interact, and that co-localization of Bicc1 with ANKS3 is not merely a non-specific consequence of any SAM domain or ANK repeat protein overexpression.
Figure 5. Cytoplasmic Clustering of Bicc1 Is Inhibited by ANKS3 C-Terminal Domain and Rescued by ANKS6.
(A) Immunofluorescent staining of individually transfected HA-Bicc1, ANKS3-Flag, ANKS3ΔCter-Flag or v5-ANKS6.
(B and C) Immunofluorescent staining of co-transfected ANKS3-Flag and v5-ANKS6 stained (B) together with anti-Flag and anti-ANKS6 antibodies or (C) only one at a time with anti-Flag or anti-v5 antibodies.
(D and E) Co-immunostaining of HA-Bicc1 with (D) v5-ANKS6 or ANKS3-Flag or with (E) C-terminally truncated ANKS3ΔCter-Flag. Bottom row in (D) shows enlargement of the boxed area in the merge panel.
(F) Co-localization of HA-Bicc1 together with ANKS3-Flag and v5-ANKS6. Scale bars, 5 μm.
To assess whether Bicc1-ANKS3 foci can be influenced by ANKS6 or vice versa, we co-transfected all three proteins. Double immunostaining of HA-Bicc1 and ANKS3-Flag or HABicc1 and v5-ANKS6 revealed that the presence of v5-ANKS6 suppressed the scattering of Bicc1 foci by ANKS3-Flag and instead induced the formation of even larger cytoplasmic domains harboring all three proteins (Figure 5F). Interestingly, ANKS3-Flag was specifically enriched at the periphery but not in the center of such foci, whereas HA-Bicc1 and v5-ANKS6 were homogeneously distributed. Besides confirming the conclusion of the co-immunoprecipitation experiments showing that ANKS6 is linked to Bicc1 by ANKS3, this result shows that ANKS6 recruitment alters the topology of macromolecular Bicc1-ANKS3 complexes in a non-random, consistent fashion.
The Bicc1-ANKS3-ANKS6 Network Does Not Rely Exclusively on SAM-SAM Interfaces
To evaluate the contribution of the Bicc1 SAM domain to ANKS3 or ANKS6 interactions, we conducted co-immunoprecipitation assays using truncated Bicc1ΔSAM or the polymerization mutant Bicc1 mut-D where residues D913, K915 and E916 (mouse Bicc1 numbering) are substituted by alanines to disrupt the ML and EH surfaces of the SAM-SAM interface (Rothé et al., 2015) (Figure S3A). Surprisingly, both mutants still co-immunoprecipitated ANKS3 alone or together with ANKS6 (Figure S3B). In parallel, we evaluated the potential of ANKS3 and ANKS6 to cluster hemagglutinin (HA)-tagged Bicc1 mut-D or Bicc1ΔSAM in cytoplasmic foci. Both Bicc1 mut-D and Bicc1ΔSAM are unable to self-polymerize in large cytoplasmic clusters on their own (Maisonneuve et al., 2009; Rothé et al., 2015). Their cytoplasmic localization also remained diffuse in the presence of ANKS3. However, when co-expressed together with ANKS6, ANKS3 significantly rescued the localization of HA-Bicc1ΔSAM or HA-Bicc1 mut-D in cytoplasmic foci (Figures S3C and S3D). These results agree with our Y2H data that Bicc1 binds ANKS3 in part independently of a SAM-SAM interaction through its KH domains, and that the ANKS3 SAM-domain interfaces with the SAM domain of ANKS6 (Figures 2B, 2D, 2F, 2I, and S2). Thus, ANKS3-ANKS6 hetero-oligomers can recruit Bicc1 in part independently of its own SAM domain, whereas Bicc1 SAM-domain polymerization reinforces this network and remodels the shape of the resulting macromolecular complexes.
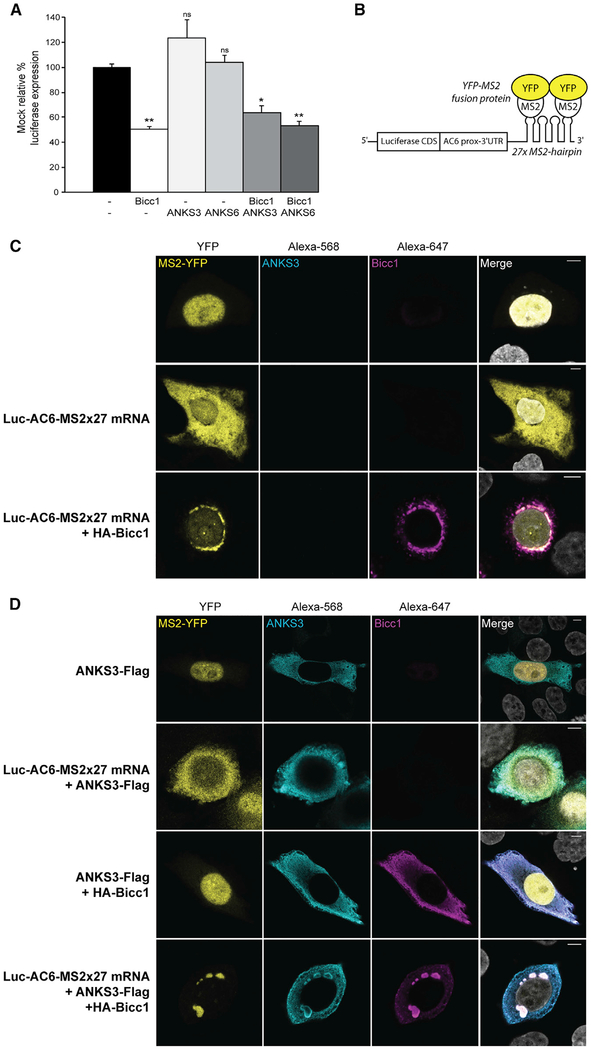
While ANKS3 and ANKS6 Do Not Affect Bicc1-Mediated Silencing of a Reporter mRNA, mRNA Binding Instead Changes the Topology of Protein Complexes
The molecular function of polymeric Bicc1 involves the cytoplasmic localization and translational repression of bound mRNA (Rothé et al., 2015). Prompted by our finding that ANKS3 and ANKS6 can modulate the topology of cytoplasmic Bicc1 clusters, we hypothesized that they regulate Bicc1-mediated silencing of the target 3′ UTR of AC6 mRNA (Piazzon et al., 2012). Surprisingly, Bicc1 repressed the Luc-AC6 3′ UTR reporter irrespective of the presence or absence of exogenous ANKS3 or ANKS6 (Figure 6A). To assess whether the mRNA itself formed a scaffold to restore large Bicc1 foci, we imaged the cytoplasmic distribution of a Luc-AC6-MS2×27 reporter mRNA by tethering fluorescent MS2-YFP fusion protein, which normally is retained in the nucleus by a nuclear localization signal (Figure 6B) (Rothé et al., 2015). As described previously, co-expression with HA-Bicc1 alone efficiently recruited the Luc-AC6-MS2×27 reporter and associated MS2-YFP to cytoplasmic HA-Bicc1 foci (Figure 6C). In the absence of Luc-AC6-MS2×27 mRNA, ANKS3-Flag with or without HA-Bicc1 as a control did not alter the nuclear localization of MS2-YFP (Figure 6D, rows 1 and 3). Co-expression with ANKS3-Flag alone also did not influence the localization of Luc-AC6-MS2×27 mRNA or vice versa (Figure 6D, row 2). However, when co-expressed together with both HA-Bicc1 and ANKS3-Flag, the reporter mRNA rescued large Bicc1 foci despite the presence of Flag-ANKS3 (Figure 6D, row 4). Moreover, Flag-ANKS3 staining was conspicuously absent at the center of such rescued Bicc1 foci and instead decorated their periphery (Figures 6D and S4), similar to what we observed when ANKS3 and Bicc1 are jointly co-expressed with ANKS6 (Figure 5F). These results suggest that RNA binding can dramatically alter the spatial configuration of macromolecular Bicc1 protein complexes.
Figure 6. Bicc1-Mediated Repression of a Reporter mRNA Is Not Affected by Exogenous ANKS3 and ANKS6.
(A) Silencing of AC6 3′UTR luciferase reporter by HA-Bicc1 in the absence or presence of ANKS3-Flag or v5-ANKS6 in transfected HEK293T cells. β-Galactosidase was co-transfected for signal normalization. Data represent mean ± SEM of 3 experiments. *p < 0.05, **p < 0.01.
(B) Principle of the MS2-YFP co-localization assay. The 3′end of the Luc-AC6 reporter mRNA was fused to 27 MS2 hairpins, which can bind the MS2 protein fusedtoYFP. CDS, codingsequence;prox, proximal.
(C and D) Localization of the Luc-AC6-MS2327 reporter mRNA and HA-Bicc1 in the absence (C) or presence (D) of ANKS3-Flag in transfected HeLa cells. The MS2-tagged mRNA was imaged by nuclear MS2-YFP fusion protein that is retained in the cytoplasm when bound to the MS2 RNA hairpins. HA-Bicc1 and ANKS3-Flag were detected by immunofluorescent staining using anti-HA or anti-Flag antibodies. Scale bars, 5 μm.
The bpk Mutation Does Not Block the Association of Bicc1 with ANKS3 or ANKS6
The bpk mouse model is a non-congenital model of ARPKD characterized by an aberrant C-terminal extension that inhibits Bicc1 polymerization and stabilization in large cytoplasmic foci (Cogswell et al., 2003; Rothé et al., 2015). To evaluate whether the bpk mutation affects interactions of Bicc1 with ANKS3 or ANKS6, we co-expressed ANKS3-Flag and v5-ANKS6 alone or in combination with bpk mutant HA-Bicc1. Similar to wild-type HA-Bicc1, the bpk mutant co-immunoprecipitated with full-length and C-terminally truncated ANKS3-Flag, and it did not impair the recruitment of ANKS6 (Figure 7A). By contrast, when co-expressed with ANKS3 and ANKS6 together in HeLa cells, Bicc1 bpk only partly accumulated in ANKS3-ANKS6 foci and partly remained diffusely localized throughout the cytoplasm (Figure 7B). Besides confirming that the ANKS6-mediated scaffolding of Bicc1-ANKS3 complexes into giant clusters is partly independent of SAM-domain polymerization, these data show that the bpk mutant phenotype is not simply caused by a lack of ANKS3 or ANKS6 binding, but possibly by failure to correctly connect such complexes in a mesh-like macromolecular network (Figure S5).
Figure 7. ANKS3 and ANKS6 Partially Rescue the Recruitment of bpk Mutant Bicc1 to Cytoplasmic Foci Despite Inhibition of Its Self-Polymerization by an Aberrantly Elongated C Terminus.
(A) Co-immunoprecipitation (IP) of ANKS3-Flag or ANKS37ΔCter-Flag and v5-ANKS6 with wild-type or bpk mutant HA-Bicc1 by anti-HA antibodies. Five percent of total HEK293T cell extracts was loaded as input. γ-Tubulin was used as negative control. Asterisk denotes an ANKS6-specific band of reduced size.
(B) Co-localization of bpk mutant HA-Bicc1 with ANKS3-Flag transfected with or without v5-ANKS6 in HeLa cells and stained with anti-HA and anti-Flag antibodies. v5-ANKS6 was unstained because anti-v5 and anti-Flag antibodies are both from mouse. Scale bars, 5 μm.
Global Analysis Revealed that SAM Domains Mainly Localize in Protein C-Terminal Regions
Since large Bicc1 foci were inhibited both by the aberrant C-terminal elongation of the bpk frameshift mutation and by ANKS3 via its long C-terminal region after the SAM domain, we queried the InterPro database (Finn et al., 2017) (https://www.ebi.ac.uk/interpro/) to systematically address whether the positioning of SAM domains is skewed in favor of long or short flanking sequences. In 178 proteins larger than 280 amino acids and containing a single SAM domain, we determined whether the center of the SAM domain resides in the N-terminal or C-terminal quartiles of the host protein, or in the middle section (from 25% to 75% of the sequence). Indeed, 68% of the SAM domains analyzed resided in the C-terminal quartile, compared with only 17% and 14% that were found in the N-terminal or middle regions (Table S1). Furthermore, in proteins where the SAM domain is positioned near the C terminus, the average number of residues after the SAM domain is only 29. This bias for SAM domains to localize near the C terminus may reduce the risk of self-aggregation of unfinished polypeptide chains during mRNA translation. Alternatively, overcrowding of the space around the SAM polymer helix by bulky N and C termini together may sterically hinder head-to-tail polymerization.
DISCUSSION
SAM domains often form polymers in vitro, but how this process or their hetero-oligomerization is regulated is unclear. Since complexes of Bicc1, ANKS3, and ANKS6 have previously been linked to polycystic kidneys, we explored the regulation of their assembly into homo- and hetero-oligomers by SAM domains and their flanking sequences and by a Bicc1 target RNA. Crystal structure determination and biophysical analysis revealed that the SAM domain of Bicc1 polymerizes in a flexible manner with itself. Alternatively, the ML and EH surfaces can also mediate co-polymerization with the SAM domain of ANKS3, but not ANKS6. Unexpectedly, reconstitution experiments in heterologous cells showed that neither ANKS3 nor ANKS6 formed macroscopic homopolymers on their own, in sharp contrast to Bicc1. However, binding to ANKS3 dispersed foci of Bicc1 polymers, whereas co-recruitment of ANKS6 by ANKS3 enlarged them. ANKS6 induced large ANKS3-ANKS6 heteropolymers that recruited Bicc1 partly independently of its SAM domain, likely through additional interfaces involving Bicc1 KH domains and ANK repeats of ANKS3. Interestingly, binding of a synthetic mRNA to Bicc1 KH domains similarly rescued the assembly of Bicc1-ANKS3 hetero-oligomers into giant foci, with a stereotypical spatial rearrangement of ANKS3 at their periphery. The potential of individual SAM domains to form homo- or heteropolymers was modulated by flanking sequences, suggesting that it cannot be directly inferred from analyzing only their interactions in isolation. Together, our observations allow us to propose biochemical rules for the self-assembly of these macro-molecular ribonucleoprotein complexes as a first approximation. Stoichiometries and the localization of hetero-oligomeric complexes likely vary in physiological contexts, but given the vast number of SAM-domain proteins, similar rules may govern the assembly of other SAM-domain-mediated hetero-oligomeric protein scaffolds.
Structure of the Bicc1 SAM Domain and Mutational Analysis of Its Interaction with ANKS3 SAM
The high-resolution crystal structure revealed that the helical SAM-domain polymer lacks radial symmetry because the ML and EH surfaces of each SAM subunit interfaced with each other in three different orientations. Interestingly, all residues validated by point mutations to be involved in polymerization made contacts in at least one of the three interfaces, but no binding interface involved all of these residues. We cannot formally rule out that such variability is an artifact linked, e.g., to the R924E mutation that we used to increase protein solubility before crystallization. However, flexible binding has previously been observed between the SAM domains of EphA2 and SHIP2, where native ML-EH interactions populate two different orientations (Lee et al., 2012). Flexibility resulting from the electrostatic nature of the ML-EH-binding interface may increase the versatility of macromolecular Bicc1 scaffolds by facilitating malleable docking of ANKS3 or other interacting proteins. Our data identify the SAM domain of ANKS3 as direct binding partner of the Bicc1-SAM. Affinity measurements and point mutations showed that the SAM:SAM interfaces in heteromeric ANKS3/Bicc1 SAM complexes have binding affinities similar to those in Bicc1-SAM (this study) and ANKS3-SAM homopolymers (Leettola et al., 2014), suggesting that they likely consist of alternating co-polymers rather than blocks of two homo-oligomers. A report that a GFP fusion of ANKS3 forms large cytoplasmic aggregates in multi-ciliated epidermis of Xenopus embryos was consistent with both scenarios but did not distinguish between them (Yakulov et al., 2015).
ANKS3 Can Disperse or Enlarge Polymeric Bicc1 Scaffolds, Depending on Its Association with ANKS6
In contrast to Bicc1- and ANKS3-SAM domains, the ANKS6-SAM previously has been shown to only interact with the EH surface of the ANKS3-SAM but not with itself (Knight et al., 2011; Leettola et al., 2014). However, interactions between full-length proteins remained to be mapped. Here, reconstitution in heterologous cells showed that neither ANKS6 nor ANKS3 alone accumulated in HMW fractions of sucrose density gradients or as macroscopic higher-order complexes in cytoplasmic foci, suggesting that they do not form long homopolymers in vivo. Thus, large cytoplasmic foci of ANKS3 described in zebrafish embryos (Yakulov et al., 2015) are likely assembled by interacting factors. Possible candidates include ANKS6 but not Bicc1, because our reconstitution analysis in HeLa cells showed that ANKS3 and ANKS6 congregate in cytoplasmic foci only if they are co-expressed (with or without Bicc1), whereas complexes of ANKS3 with Bicc1 alone were dispersed throughout the cytoplasm. In good agreement, our Y2H and co-immunoprecipitation assays showed that ANKS3 independently binds Bicc1 and ANKS6. In sharp contrast, binding of Bicc1 to ANKS6 was minimal, except in the presence of ANKS3. These results strongly suggest that ANKS6-Bicc1 complexes previously described in kidneys (Bakey et al., 2015) are stabilized by ANKS3. Previous analysis of Bicc1 and ANKS6 deletion constructs predicted that they depend on a third partner to mediate their interaction between the ANKS6-SAM and Bicc1-KH domains (Stagner et al., 2009). We propose that this partner is ANKS3. Overall, our findings suggest that only Bicc1 forms self-polymers on its own and that ANKS3 limits their enlargement, whereas co-recruitment of ANKS6 promotes it, possibly by capping the ANKS3-SAM domain so that segments of homo-polymeric Bicc1 have more chance to form inside the macromolecular complexes (Figure S5).
Giant Modular Protein Complexes of Bicc1, ANKS3, and ANKS6 Form through an Intricately Interwoven Network of Protein Interactions
The estimated size of endogenous Bicc1 foci in kidney cells and of homopolymeric Bicc1 HMW complexes in sucrose gradients is larger than ribosomes (Rothé et al., 2015). Bicc1-ANKS3-ANKS6 complexes in cytoplasmic foci and on density fractionation gradients were even larger, with no obvious constraints limiting their growth. What regulates the size of foci in renal tissue is unknown. Both bpk mutant Bicc1 and two other polymerization mutants, Bicc1 mut-D and Bicc1ΔSAM, still formed cytoplasmic foci with ANKS3 and ANKS6. However, compared with wild-type Bicc1 foci scaffolded by SAM-SAM interactions, they were smaller and surrounded by abnormally diffuse Bicc1, suggesting that complete assembly of large Bicc1-ANKS3-ANKS6 complexes requires both the scaffolding by Bicc1 SAM polymerization and additional interactions. In good agreement, our Y2H experiments suggest that ANKS3 and Bicc1 interconnect both via their SAM domains and by their ANK repeats and KH domains. In addition, the SAM domain and ANK repeats of ANKS3 strongly bound those of ANKS6, and Bicc1-KH domains also formed direct contacts with full-length Bicc1. Thus, the assembly of Bicc1 hetero-oligomers clearly involves not only SAM domains. A mesh-like structure encompassing SAM homo-and/or heteropolymers linked through additional interactions is more plausible (Figures S5G and S5H). In addition, the regulation of the relative concentrations of individual components of these hetero-oligomeric complexes, and their post-translational modifications or their sequestration in cilia or other compartments, may also play a role.
Effect of a Target mRNA on the Size and Topology of Bicc1 Protein Complexes
To our surprise, dispersal of large Bicc1 foci by ANKS3 into small ones was also suppressed in cells overexpressing an mRNA that contains the Bicc1-interacting region of the AC6 3′ UTR. The simplest explanation might be that mRNA can link small Bicc1-ANKS3 foci to each other into big ones (Figure S5I). Such a model is consistent with the fact that Bicc1 and ANKS6 are linked by ANKS3 and co-immunoprecipitate more efficiently before than after RNase A treatment (Stagner et al., 2009). The pivot used by mRNA to enlarge Bicc1-ANKS3 complexes likely involves the Bicc1 KH domains. Since Bicc1-KH domains can also directly contact ANKS3 and full-length Bicc1, future studies should investigate whether such contacts affect mRNA binding or vice versa. Protein scaffolding functions have been reported for long non-coding RNAs, which bring molecules in close proximity to enhance their interactions and functions in crowded cellular environments. However, to our knowledge no scaffolding functions have been described for mRNAs. Thus, besides being a target, the mRNA might serve to regulate associated protein complexes.
Modulation of SAM Polymerization by Flanking Sequences
Previously we have shown that Bicc1 SAM domain self-polymerization was blocked by a frameshift mutation in Bicc1bpk that aberrantly elongates the C terminus by 149 amino acids (Rothé et al., 2015). Similarly, clustering by a SAM domain is inhibited in the unrelated protein Polyhomeotic upon fusion to a C-terminal GFP tag (Isono et al., 2013). Here, the deletion of the C-terminal domain that flanks the SAM domain of ANKS3 was not sufficient to induce its self-polymerization, but enabled it to form HMW complexes with Bicc1. According to structure models, helical SAM polymers are expected to display N- and C-terminal flanking sequences at their periphery (for review see Qiao and Bowie, 2005), suggesting that they may impose important steric constraints if their size or shape forces them to clash with each other. Here, our systematic analysis of 180 human proteins revealed that their SAM domains reside in the C-terminal quartile in 68% of all cases, consistent with a possible link between C-terminal positioning and the ability of a given SAM domain to polymerize. Although few SAM domains have been confirmed to polymerize in the context of their full-length proteins, some form self-polymers despite their positioning close to the N terminus, e.g., in SLP-76 (Liu et al., 2013). Nevertheless, the distribution bias in SAM location inside the protein sequence points to a selection mechanism favoring SAM domains close to C termini.
Among structural domains frequently associated with SAM domains in 178 proteins analyzed (Table S1), ANK repeats are found for 18 entries (10%), whereas KH domains are only found in Bicc1. Of note, in every case ANK repeats are upstream of the SAM domain and localize in the N-terminal half of the protein, suggesting that this combination is particularly suited for a molecular scaffolding function. Indeed, while the SAM domain is a powerful domain for polymerizing and concentrating proteins, ANK repeats are versatile platforms for the recruitment of other interacting partners (for review see Mosavi et al., 2004). A recent example is the poly(ADP-ribose) polymerase Tankyrase and its role in Wnt signaling, whereby ANK repeats serve as a docking site for target proteins and SAM-dependent polymerization potentiates their modification by ADP-ribose (Mariotti et al., 2016; Riccio et al., 2016).
Are Bicc1-ANKS3-ANKS6 Scaffolds Acting as Modular Signaling Platforms?
Recent findings reported that three SAM proteins, namely Sfmbt, Scm, and Ph, exhibit different polymerization properties and interact together in a hierarchic manner to scaffold the assembly of Polycomb (PcG) protein complexes on the chromatin and to induce transcriptional repression (Frey et al., 2016). Whether macromolecular Bicc1-ANKS3-ANKS6 complexes act as modular scaffolds to similarly regulate specific signaling effectors remains to be determined. In the cAMP/PKA signaling pathway, SAM-mediated polymerization is required for translational repression of AC6 and PKIa mRNAs, but a role for ANKS3 or ANKS6 in regulating Bicc1-mediated mRNA silencing has not been found. Polymerization also increases Bicc1 stability (Rothé et al., 2015), which is important for curbing mTORC1 and canonical Wnt signaling (Maisonneuve et al., 2009; Shillingford et al., 2012). However, even a polymerization-defective mutant Bicc1 inhibits canonical Wnt signals if it is expressed at normal levels (Rothé et al., 2015). It will be interesting to investigate in future studies whether a scaffolding function affects the activation of the kinase NEK8 by ANKS6 and its effect on YAP/TAZ signaling (Czarnecki et al., 2015; Hoff et al., 2013), or the cytoplasmic retention of NEK7 by ANKS3 (Ramachandran et al., 2015).
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-HA (produced in rabbit) | Sigma | H6908; RRID: AB_260070 |
| Monoclonal anti-v5 (produced in mouse) | ThermoFisher | MA5-15253; RRID: AB_10977225 |
| Monoclonal anti-FLAG® M2 (produced in mouse) | Sigma | F3165; RRID: AB_259529 |
| Polyclonal anti-ANKS6 (produced in rabbit) | Sigma | HPA008355; RRID: AB_2058277 |
| Monoclonal anti-HA-Agarose antibody (produced in mouse) | Sigma | A2095; RRID: AB_257974 |
| Anti-FLAG® M2 affinity gel (produced in mouse) | Sigma | A2220; RRID: AB_10063035 |
| Bacterial and Virus Strains | ||
| Rosetta(DE3) pLysS cells | Novagen | 70956 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-Amino-1,2,4-triazole ≥95% (TLC) | Sigma | A8056 |
| RunBlue Native Run Buffer | Expedeon | NXB61500 |
| Critical Commercial Assays | ||
| Ni-NTA Superflow agarose | Qiagen | 30450 |
| Superdex 200 10/300 GL column | GE Healthcare | 28990944 |
| WTC-030S5 analytical size-exclusion column | Wyatt Technology Co. | N/A |
| Deposited Data | ||
| Crystal structure of the SeMET BICC1 SAM Domain R924E mutant | PDB | 4RQM |
| Crystal structure of the native BICC1 SAM Domain R924E mutant | PDB | 4RQN |
| Experimental Models: Cell Lines | ||
| HEK293T | N/A | N/A |
| HeLa | N/A | N/A |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae strain CG1945 | IMOPA, Pr. B. Charpentier | N/A |
| S. cerevisiae strain Y187 | IMOPA, Pr. B. Charpentier | N/A |
| Oligonucleotides | ||
| See Table S2 for primers used for PCR amplification | this paper | N/A |
| Recombinant DNA | ||
| pCMV-SPORT6::HA-Bicc1 | Maisonneuve et al., 2009 | N/A |
| pCS+::AC6–3’UTRprox-MS2×27 | Rothé et al., 2015 | N/A |
| pMS2-YFP | Addgene | 27122 |
| pcDNA6::V5-ANKS6 | Dr. S. Lienkamp, Hoff et al., 2013 | N/A |
| pCMV6-Entry::ANKS3-Flag, | Origene | RC223862 |
| pCMV6-Entry::ANKS3DCter-Flag, | this paper | N/A |
| pACT2::BICC1-FL | this paper | N/A |
| pACT2::BICC1-KH | this paper | N/A |
| pACT2::BICC1-IVS | this paper | N/A |
| pACT2::BICC1-SAM | this paper | N/A |
| pACT2::BICC1-ANKS3-FL | this paper | N/A |
| pACT2::BICC1-ANKS3-ANK | this paper | N/A |
| pACT2::BICC1-ANKS3-SAM | this paper | N/A |
| pACT2::BICC1-ANKS6-FL | this paper | N/A |
| pACT2::BICC1-ANKS6-ANK | this paper | N/A |
| pACT2::BICC1-ANKS6-SAM | this paper | N/A |
| pHis-SUMO::BICC1 SAM | this paper | N/A |
| pH is-SUMO::ANKS3 SAM | Leettola et al., 2014 | N/A |
| pBAD-HisA-negGFP::B ICC1 SAM | this paper | N/A |
| pBAD-HisA-negGFP::ANKS3 SAM | Leettola et al., 2014 | N/A |
| pBAD-HisA-negGFP::ANKS6 SAM | Leettola et al., 2014 | N/A |
| Software and Algorithms | ||
| ASTRA | Wyatt Technology Co. | http://www.wyatt.com/products/software/astra.html |
| Biacore T100 Evaluation | GE Healthcare | N/A |
| XDS | Kabsch, 2010 | N/A |
| PROCHECK | Laskowski et al., 1993 | https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/index.html |
| PyMOL | Schrodinger, LLC | http://www.pymol.org |
| PISA | Krissinel and Henrick, 2007 | http://www.ccp4.ac.uk/pisa/ |
| VERIFY3D | Bowie et al., 1991 | http://services.mbi.ucla.edu/Verify_3D/ |
| ERRAT | Colovos and Yeates, 1993 | http://services.mbi.ucla.edu/ERRAT/ |
| SHELX | Pape and Schneider, 2004 | http://shelx.uni-ac.gwdg.de/tutorial/english/verf.htm |
| HKL2MAP | Pape and Schneider, 2004 | http://webapps.embl-hamburg.de/hkl2map/ |
| CCP4 suite | Winn et al., 2011 | http://www.ccp4.ac.uk/ |
| PHENIX | Adams et al., 2010 | N/A |
| BUSTER | Bricogne et al., 2011 | https://sbgrid.org/software/titles/buster |
| PHASER | McCoy et al., 2007 | https://www.phenix-online.org/documentation/reference/phaser.html |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| IMARIS | Bitplane | http://www.bitplane.com/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel B. Constam (daniel.constam@epfl.ch).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Standard S. cerevisiae growth and handling techniques were employed to culture the yeast two-hybrid strains CG1945 and Y187. Non-transformed cells were grown in Yeast extract Peptone Dextrose medium (YPD). After transformation, cells were selected and grown in Standard Minimal Medium (SD) lacking amino acids used for the selection.
Human HEK293T and HeLa cell lines were cultured in Dulbecco modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum (FBS; Sigma), glutamine (1%; Invitrogen), and gentamicin (1%; Invitrogen). Plasmids were transfected using the jetPEI transfection reagent (Polyplus Transfection) according to the manufacturer’s instructions.
METHOD DETAILS
Human BICC1 has been used for X-ray structure, biophysics and Y2H. Mouse Bicc1 has been used in co-immunoprecipitation, sucrose gradient and imaging experiments to allow direct comparisons with bpk mutant Bicc1, and with two other mouse mutants (mutD and ΔSAM) that were previously characterized (Maisonneuve et al., 2009; Piazzon et al., 2012; Rothé et al., 2015). Human and mouse Bicc1 are 93% identical. In their KH and SAM domains, sequence identity even increases to 97% and 100%, respectively, and proteins from both species behaved similarly in all experiments analyzed in this study.
Plasmids and cDNA Cloning
SAM domain fusions of human Bicc1 (residues 870–939), ANKS3 (residues 421–490), and ANKS6 (residues 771–840) with negGFP or hexahistidine small ubiquitin-like modifier (SUMO) tags were cloned as described previously (Knight et al., 2011; Leettola et al., 2014). The Quickchange method (Agilent) was used for site-directed mutagenesis. Mammalian expression plasmids pCMV-SPORT6:: HA-Bicc1, pCS+::AC6–3’UTRprox-MS2×27 and pcDNA6::V5-ANKS6 (Hoff et al., 2013; Rothé et al., 2015) have been described previously. ANKS3 cDNA (NM_133450), expressed from pCMV6-Entry and fused to C-terminal Flag tag, was from OriGene (clone ID: RC223862). For pCMV6-Entry::ANKS3ΔCter-Flag, the coding sequence for amino acids 1 to 490 was amplified by PCR and subcloned between BamHI and MluI sites of pCMV6-Entry. For Y2H, Bicc1, ANKS3 and ANKS6 coding fragments were amplified by PCR and subcloned in pACT2 and pGBKT7 plasmids using BamHI-XhoI and BamHI-SalI restriction sites, respectively. Primers used for PCR amplification are listed in Table S2.
Protein Expression and Purification Protein Expression and Purification
SAM domai2ns used for structural and biophysical analysis were expressed as pHis-SUMO constructs in Rosetta(DE3) pLysS cells (Novagen) and processed as described (Leettola et al., 2014). Briefly, lysate supernatant from harvested cells, clarified by centrifugation at 13,200g for 20 min at 4°C and supplemented with 10 mM imidazole, was bound to Ni-NTA Superflow agarose (Qiagen). Loaded columns were washed twice with 8 volumes of lysis buffer (20 mM NaHPO4 pH 8, 0.5 M NaCl) containing 15 mM or 20 mM imidazole, respectively, prior to elution in lysis buffer containing 250 mM imidazole. Eluted protein was diluted to 5 mg/ml and dialyzed with 20 mM Tris pH 8, 0.5 M NaCl. To remove His6-SUMO tags, dialyzed proteins were digested with SUMO protease 1 (ULP1) catalytic domain at a 50:1 protein:protease molar ratio for 16 hrs at 4°C and passed back over Ni-NTA resin (Malakhov et al., 2004). Unbound proteins in the flow-thru were further purified as described in the table below. ANKS3 and ANKS6 SAM domain constructs were produced as described previously (Leettola et al., 2014). Selenomethionine (SeMet) labeled Bicc1 R924E was produced using an adapted version of a previously described protocol (Van Duyne et al., 1993). An overnight culture of Rosetta(DE3)pLysS cells transformed with pHis-SUMO Bicc1 R924E was grown in M9 minimal media supplemented with kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml). 20 ml of overnight culture was used to inoculate each of three 1L flasks of M9 minimal media and cells were grown at 37°C with shaking until an OD600 of 0.55 was reached, at which point SeMet (60 mg), Lys, Phe, and Thr (100 mg each) and Ile, Leu, Val (50 mg each) were added. Cells were incubated for another 15 min at 37°C, then cooled to 18°C, induced with 1 mM IPTG and expressed overnight at 18°C with shaking. Harvested cells were resuspended in 30 ml of degassed lysis buffer (20 mM Tris.HCl pH 7.5, 0.5 M NaCl, 10 mM βME) supplemented with 1 mM PMSF and 20 μg/ml DNase1 and lysed as described previously (Leettola et al., 2014). Purified proteins were polished as summarized in Table S3.
Crystallization and Structure Determination
We used multi-wavelength anomalous dispersion (MAD) to solve a selenomethionine (SeMet)-replaced Bicc1 SAM R924E mutant to 1.75Å resolution. We also crystallized a native R924E mutant which had a slightly more compact unit cell and solved this structure to 2.00Å resolution by molecular replacement. Both the native and SeMet constructs crystallized in space group P212121 with 3 molecules in the asymmetric unit. The coordinates have been deposited in the PDB with accession codes 4RQM and 4RQN. Crystallization trials were initiated at the Macromolecular Crystallization Core Technology Center at UCLA using commercially available screens and a Mosquito crystallization robot (TTP Labtech) to set up hanging drop vapor diffusion experiments. SeMet derivative crystals were grown by hanging drop vapor diffusion in 1 μl drops prepared by a Mosquito crystallization robot (TTP Labtech). Bicc1-SAM R924E SeMet at 30.3 mg/mL was mixed with well solution (100 mM MES pH 7, 35% PEG-550 MME, 5mM ZnSO4) in a 2:1 protein:reservoir ratio. Boulder-shaped crystals grown at room temperature for 2.5 weeks were cryoprotected in the same solution supplemented with an additional 10% PEG-550 MME. Single crystals were mounted with CrystalCap HT Cryoloops (Hampton Research, Aliso Viejo). Because of crystal decay, the MAD data sets were collected using 2 crystals cryo-cooled to 100 K at the Advanced Photon Source (Argonne National Laboratory), APS-NECAT beamline 24-ID-C on a DECTRIS-PILATUS 6 M detector. The first crystal was used to collect at the peak (0.9791Å) and contained 600 0.2o oscillation frames with diffraction extended to 1.75Å. The second crystal was used to collect the inflection and high remote datasets (0.9793Å and 0.9714Å) at 2.4Å resolution. Each data set contained 720 0.2o oscillation frames. Datasets were processed using XDS (Kabsch, 2010). Phasing was accomplished by MAD using the HKL2MAP interface and SHELX programs (Pape and Schneider, 2004). Three selenomethionines were located with SHELXD and used for phasing in SHELXE. Density modification and model building were performed using DM and BUCCANEER in the CCP4 suite (Winn et al., 2011). The structure was refined in PHENIX (Adams et al., 2010) and BUSTER (Bricogne et al., 2011) using individual sites, individual atomic displacement parameters, non-crystallographic symmetry, and TLS parameterization with 3 TLS groups per chain. TLS groups were chosen by inspection of the structural model and each chain was divided into 3 groups: 1) helices 1–2; 2) helices 3–4; and 3) helix 5. After each round of refinement, the model was visually inspected and problem areas rebuilt in COOT (Emsley et al., 2010). Data collection and refinement statistics are reported in Table 1. The coordinates have been deposited in the PDB with accession code 4RQM. Native crystals were grown by hanging drop vapor diffusion in 500 nl drops prepared by a Mosquito crystallization robot (TTP Labtech). Bicc1-SAM R924E at 29.8 mg/ml was mixed with well solution (100mM MES pH 6.5, 30% PEG-550 MME, 5mM ZnSO4) in a 2:1 protein:reservoir ratio. Boulder-shaped crystals grew at room temperature over 10 days and were cryoprotected using well solution supplemented with an additional 10% PEG-550 MME. A high resolution data set was collected on a single native crystal cryo-cooled to 100 K at the Advanced Photon Source (Argonne National Laboratory), APS-NECAT beamline 24-ID-C on a DECTRIS-PILATUS 6 M detector. A data set containing 720 0.5° oscillation frames was collected from a single large crystal at a wavelength of 0.97950Å and processed using XDS (Kabsch, 2010). Molecular replacement was performed using PHASER with chain C of the BICC1-SAM R924E SeMet structure as a search model and searching for 3 copies in the asymmetric unit (McCoy et al., 2007). The model was refined using PHENIX with TLS parameterization including individual sites, individual atomic displacement parameters, and noncrystallographic symmetry (Adams et al., 2010). Data collection and refinement statistics are reported in Table 1.
Structure and Sequence Analysis
ClustalW2 was used to perform multiple sequence alignments (Larkin et al., 2007). PROCHECK (Laskowski et al., 1993), ERRAT (Colovos and Yeates, 1993), and VERIFY3D (Bowie et al., 1991) algorithms were used to validate the final structural models. Figures were prepared in PyMOL (DeLano, 2010). Surface electrostatics were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) plugin in Pymol, and all surfaces were contoured at ±1 kT/e (Baker et al., 2001). Analysis of protein-protein binding interfaces was accomplished using PISA (Krissinel and Henrick, 2007).
negGFP Native Gel Binding Assays
Native gel binding assays were performed as previously described (Leettola et al., 2014). Lysates of ARI814 cells expressing negGFP-human-SAM fusions were loaded on 20% RunBlue 12-well Native gels in RunBlue Native Run Buffer (Expedeon) and developed at 90V for 16 hrs at 4°C. Gels were visualized with a Bio-Rad Molecular Imager FX Pro-Plus and a Bio-Rad Molecular Imager PharosFX using an excitation wavelength of 488 nm and an emission wavelength of 510 nm. To monitor hetero-SAM interactions, equal amounts of each protein (as determined by fluorescence) were mixed and allowed to equilibrate at 4°C for 4 hrs prior to gel loading.
Analytical Size-Exclusion Chromatography
The interaction between the recombinant Bicc1 and ANKS3 SAM domains was assessed by size-exclusion chromatography. Size Exclusion Chromatography with Multi-Angle Light Scattering analyses were performed as previously described (Leettola et al., 2014). For analysis of the Bicc1-EH/ANKS3-ML binding, 250 ul of protein in 20 mM HEPES pH 7.5, 0.15 M NaCl, 1 mM DTT was loaded on a Superdex 200 10/300 GL column (GE) equilibrated in the same buffer at a flow rate of 0.4 ml/min. Proteins were loaded as follows: Bicc1 E898K was loaded at 2 mg/ml; ANKS3 F472E and BICC1 D911K at 4mg/ml; the 1:1 molar ratio mix of Bicc1 E898K + ANKS3 F472E at 4mg/ml; and the 1:1 molar ratio mix of Bicc1 D911K + ANKS3 F472E at 8 mg/mL. Elution was monitored by absorbance at 280 nm.
Analytical Size-Exclusion Chromatography and SEC-MALS
The interaction between the recombinant Bicc1 and ANKS3 SAM domains was assessed by size-exclusion chromatography. Size Exclusion Chromatography with Multi-Angle Light Scattering analyses were performed as previously described (Leettola et al., 2014). Protein samples at 10 mg/ml were analysed by SEC-MALS. 50 μl of Bicc1 F920E + ANKS3 I455E and 100μL of Bicc1 R924E + ANKS3 I455E were loaded onto a WTC-030S5 analytical size-exclusion column (Wyatt Technology Co.) equilibrated with 0.15M NaCl in 20 mM Tris HCl pH 8, 2mM βME using an AKTA purifier (GE) at 0.7 ml/min on a miniDAWN TREOS (Wyatt Technology Co.). Molecular weights and the monodispersity of protein peaks were analysed using ASTRA software (Wyatt Technology Co.).
Surface Plasmon Resonance
Experiments were performed at 21°C in 20 mM HEPES pH 7.5, 0.04% IGEPAL CA-630, 0.15 M NaCl (+1mM DTT for ANKS3 samples only) using a Biacore T100 (GE). To determine the binding affinity of the Bicc1-SAM/Bicc1-SAM interface, Bicc1-SAM R924E was immobilized on a Biacore CM5 chip (GE) via EDC/NHS crosslinking. Bicc1-SAM mutants E898K and D911K at varying concentrations were passed over the chip and equilibrium binding levels were measured. As controls, varying concentrations of the Bicc1 SAM mutants F920E and K925E (both with defective EH interfaces) and the Bicc1 double mutants E898K/F920E and D911K/F920E (each with defective ML and EH interfaces) at 10 μM were passed over the chip. To determine the binding affinity of the Bicc1-SAM ML/ANKS3-SAM EH interface, ANKS3 SAM I455E at varying concentrations was passed over the Bicc1-SAM R924E conjugated chip and equilibrium binding levels were measured. To determine the binding affinity of the Bicc1-SAM EH/ANKS3-SAM ML interface, ANKS3-SAM F472E was immobilized on a Biacore CM5 chip (GE) via EDC/NHS crosslinking. Bicc1-SAM mutants E898K and D911K at varying concentrations were passed over the chip and equilibrium binding levels were measured. As a control, the Bicc1 double mutants E898K/F920E and D91K/F920E were passed over the ANKS3-SAM F472E-conjugated chip. All binding data represent triplicate values and were fit to a 1:1 steady-state model using Biacore T100 Evaluation software. Special handling of ANKS3-SAM I455E to maintain protein stability was as described previously (Leettola et al., 2014).
Yeast Two-Hybrid Assay
Protein-protein interaction by yeast two-hybrid assays were performed as described (Rothé et al., 2014). To assess interactions between Bicc1, ANKS3, ANKS6 and their respective domains, they were each fused to the DNA-binding domain of the GAL4 transcription factor (GAL4-BD) in the pGBKT7 plasmid (Clontech) as bait proteins. In parallel, we fused them each to the activation domain of the GAL4 transcription factor (GAL4-AD) in the pACT2 plasmid (Clontech) as prey proteins. The reporter gene used in this study is the HIS3 gene required for histidine biosynthesis. To monitor bait and prey interactions, appropriate pACT2 (LEU2) and pGBKT7 (TRP1) plasmids were transformed into haploid cells from strain CG1945 (mat a; ura3–52, his3–200, ade2–101, lys2–801, trp1–901, leu2–3, 112, gal4–542, gal80–538, cyhr2, LYS2::GAL1UAS-GAL1TATA-HIS3, URA3::GAL417-mers(x3)-CYC1TATALacZ) and strain Y187 (mat α; gal4, gal80, ade2–101, his3–200, leu2–3,112, lys2–801, trp1–901, ura3–52, URA3::Gal1UAS GAL1TATA LacZ), respectively, using the lithium acetate method. After crossing on YPD medium, diploid cells were selected on media suitable for double selection (Leu-, Trp-) and then plated on media suitable for triple selection (Leu-, Trp-, His-). Where indicated, 3-Amino-1, 2, 4-triazol (3-AT) was added as a competitive inhibitor of histidine synthesis to evaluate the strength of the interactions (Rothé et al., 2014). Growth was assessed after three days of incubation at 30°C.
Co-immunoprecipitation
Protein co-immunoprecipitation from transfected HEK293T cells were performed as described (Rothé et al., 2015). HEK293T cells were transfected with 2 μg of each expression plasmids in 1×10 cm dish per condition. 24 hrs after transfection, cells were washed with PBS and proteins were extracted with lysis buffer containing 10 mM Tris.HCl pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.05% NP-40, 5% glycerol, 1 mM DTT and 1× protease inhibitor cocktail (Roche). After sonication and centrifugation at 10’000 × g for 10 min, supernatants were incubated on a rotating wheel for 2 hrs at 4°C with anti-HA agarose antibody or anti-FLAG® M2 affinity gel (Sigma). Beads were washed three times with 1 ml of 10 mM Tris.HCl pH 7.4, 200 mM NaCl, 2 mM MgCl2, 0.1% NP-40, 1 mM DTT washing buffer and resuspended in Laemmli buffer. Elutions were fractionated on SDS-PAGE gels and analyzed by Western blotting.
Sucrose Gradient Fractionation Assays
HEK293T cell extracts were fractionated on continuous 15–60% sucrose gradients as described in (Rothé et al., 2015). HEK293T cells were transfected in 10 cm dishes, using 1 plate per condition (2 μg DNA/plate). Cell extracts were prepared as mentioned above for immunoprecipitation assays. Continuous 15–60 % sucrose gradients were prepared manually by layering and passive diffusion of sucrose solutions prepared in buffer 20 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.05 % NP-40. Identical volumes of cell extracts were fractionated at 4°C by centrifugation at 100000 g for 3 hrs. Fractions were recovered manually from the top, fractionated on SDS-PAGE and analyzed by Western-blot. Error bars represent the standard error or the mean (SEM).
Indirect Immunofluorescence Analyses
Immunostaining in HeLa cells were performed as described in (Rothé et al., 2015). For immunostaining, HeLa cells were transfected with protein and RNA expression plasmids (1 μg DNA each) in 6-well plate. After 24 hrs, cells were splitted and grown in 24-well plate on sterile coverslips. Forty-eight hours after transfection, cells were fixed for 10 minutes at −20°C in methanol, washed with PBS. Coverslips were incubated 1 hr at RT for blocking in PBS containing 1% BSA, and then for 2 hrs at RT in blocking buffer containing the primary antibodies anti-HA (1/500, rabbit, Sigma), anti-v5 (1/1000, mouse, Termo Fisher), anti-Flag-M2 (1/500, mouse, Sigma) or anti-ANKS6 (1/500, Rabbit, Sigma) antibodies. After washes in PBS, the secondary antibodies anti-mouse Alexa 568 and anti-rabbit Alexa 647, were incubated in blocking buffer for 1 hr at RT in presence of DAPI (1/10000). Pictures were acquired on a Zeiss LSM700 confocal microscope.
Luciferase Assays
HEK293T cells were plated in 24-well plates. After 12 hrs, quadruplicate samples were transfected with the indicated plasmids (1× Dose: 0.1 μg/well) and with a lacZ expression vector (0.05 μg/well) using jetPEI (Polyplus Transfection). 36 hrs after transfection, cells extracts were prepared in buffer 25 mM Tris-phosphate, pH 7.8, 2 mM DTT, 2 mM CDTA, 10% glycerol, 0.5 % Triton X-100. The measurements of luciferase expression levels were carried out using 20-fold diluted extracts and luminescent counts were normalized to β-galactosidase activity. Results represent mean values of 3 independent experiments performed in quadruplicates, error bars show standard error of the mean. Student’s t test was used to calculate p values.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were used in Figures 6A and 4B–4E. Statistical details of these experiments are described in the figure legends and in the Method Details section. For luciferase assays, results represent mean values of 3 independent experiments. For each independent experiment, luciferase activities were measured in quadruplicates transfected individually for each condition tested. The error bars show standard error of the mean (SEM). Student’s t test was used to calculate P values, with P ≤ 0.05 or P ≤ 0.01 represented by one or two asterisks, respectively. For density gradient fractionation, the graphs show the percentage of protein per fraction relative to the cumulated signal of all fractions analyzed. Results represent mean values of at least 2 independent experiments. Error bars show SEMs.
DATA AND SOFTWARE AVAILABILITY
The coordinates of the SeMet and native Bicc1 SAM R924E polymer X-ray structures have been deposited in the PDB with accession codes 4RQM and 4RQN, respectively. The dataset on localization of SAM domains in human protein sequences is reported in Table S1. The human SAM protein sequences were retrieved from InterPro database (https://www.ebi.ac.uk/interpro/). The dataset encompasses 178 proteins larger than 280 amino acids and containing a single SAM domain. SAM, ANK and KH domains positioning is given in % of the total protein length.
Supplementary Material
Highlights.
Crystal structure of the Bicc1 SAM homopolymer reveals an unusually high flexibility
ANKS3, through its C-terminal domain, disperses polymericBicc1 foci
ANKS6 associates with Bicc1 via ANKS3 to rescue macromolecular complexes assembly
Bicc1, ANKS3, and ANKS6 associate in a mesh-like network through multiple interfaces
ACKNOWLEDGMENTS
We are grateful to Dr. Arne Seitz (EPFL) and the EPFL technical staff for assistance with imaging. We thank Brendan Amer for assistance with SEC-MALS. We thank Mike Collazo and Mike Sawaya at the UCLA-DOE X-ray Crystallization and Crystallography Core Facilities, which are supported by DOE grant DE-FC02–02ER63421. We thank M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov, and F. Murphy at NECAT beamlines 24-ID at APS, which are funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by an NIH-ORIP HEI grant (S10 RR029205). Use of the APS is supported by DOE under contract no. DE-AC02–06CH11357. This work was supported by grants from Gebert Rüf Stiftung (Rare Diseases grant GRS-051/13) and the Stiftung für Wissenschaftliche Forschung (Zürich) to D.B.C., NIH grant 5R01DK100482 to J.U.B., and a Ruth L. Kirschstein National Research Service award (GM007185) to C.N.L.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and three tables and can be found with this article online at https://doi.org/10.1016/j.str.2017.12.002.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakey Z, Bihoreau MT, Piedagnel R, Delestré L, Arnould C, de Villiers Ad, Devuyst O, Hoffmann S, Ronco P, Gauguier D, et al. (2015). The SAM domain of ANKS6 has different interacting partners and mutations can induce different cystic phenotypes. Kidney Int. 88, 299–310. [DOI] [PubMed] [Google Scholar]
- Bienz M (2014). Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem. Sci 39, 487–495. [DOI] [PubMed] [Google Scholar]
- Bowie J, Lüthy R, and Eisenberg D (1991). A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart O, Vonrhein C, et al. (2011). BUSTER v.2.11.2 (Global Phasing; ). [Google Scholar]
- Brown JH, Bihoreau MT, Hoffmann S, Kränzlin B, Tychinskaya I, Obermüller N, Podlich D, Boehn SN, Kaisaki PJ, Megel N, et al. (2005). Missense mutation in sterile motif of novel protein SamCystin is associated with polycystic kidney disease in (cy/+) rat. J. Am. Soc. Nephrol 16, 3517–3526. [DOI] [PubMed] [Google Scholar]
- Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, and Bryda EC (2003). Positional cloning of jcpk/bpk locus of the mouse. Mamm. Genome 14, 242–249. [DOI] [PubMed] [Google Scholar]
- Colovos C, and Yeates TO (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Gabriel GC, Manning DK, Sergeev M, Lemke K, Klena NT, Liu X, Chen Y, Li Y, San Agustin JT, et al. (2015). ANKS6 is the critical activator of NEK8 kinase in embryonic situs determination and organ patterning. Nat. Commun 6, 6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2010). The PyMOL Molecular Graphics System, Version 1.3r1 (Schrödinger, LLC; ). [Google Scholar]
- Delestré L, Bakey Z, Prado C, Hoffmann S, Bihoreau MT, Lelongt B, and Gauguier D (2015). ANKS3 co-localises with ANKS6 in mouse renal cilia and is associated with vasopressin signaling and apoptosis in vivo in mice. PLoS One 10, e0136781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, et al. (2017). InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey F, Sheahan T, Finkl K, Stoehr G, Mann M, Benda C, and Müller J(2016). Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev. 30, 1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, and Katsanis N (2011). Ciliopathies. N. Engl. J. Med 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff S, Halbritter J, Epting D, Frank V, Nguyen T-MT, van Reeuwijk J, Boehlke C, Schell C, Yasunaga T, Helmstädter M, et al. (2013). ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet 45, 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis D, Monti M, Renda M, van Koppen A, Tammaro R, Chiaravalli M, Cozzolino F, Pignata P, Crina C, Pucci P, et al. (2017). The centrosomal OFD1 protein interacts with the translation machinery and regulates the synthesis of specific targets. Sci. Rep 7, 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, and Koseki H (2013). SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 26, 565–577. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, and Diederichs K (2012). Linking crystallographic model and data quality. Science 336, 1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MJ, Leettola C, Gingery M, Li H, and Bowie JU (2011). A human sterile alpha motif domain polymerizome. Protein Sci. 20, 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus MR-C, Clauin S, Pfister Y, Di Maïo M, Ulinski T, Constam D, Bellanné-Chantelot C, and Grapin-Botton A (2012). Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum. Mutat 33, 86–90. [DOI] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, and Thornton JM (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr 26, 283–291. [Google Scholar]
- Lee HJ, Hota PK, Chugha P, Guo H, Miao H, Zhang L, Kim SJ, Stetzik L, Wang BC, and Buck M (2012). NMR structure of a heterodimeric SAM: SAM complex: characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2. Structure 20, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leettola CN, Knight MJ, Cascio D, Hoffman S, and Bowie JU (2014). Characterization of the SAM domain of the PKD-related protein ANKS6 and its interaction with ANKS3. BMC Struct. Biol 14, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Thaker YR, Stagg L, Schneider H, Ladbury JE, and Rudd CE (2013). SLP-76 Sterile? Motif (SAM) and individual H5? Helix mediate oligomer formation for microclusters and T-cell activation. J. Biol. Chem 288, 29539–29549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, and Constam DB (2009). Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136, 3019–3030. [DOI] [PubMed] [Google Scholar]
- Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, and Butt TR (2004). SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5, 75–86. [DOI] [PubMed] [Google Scholar]
- Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F, Morris E, and Guettler S (2016). Tankyrase requires SAM domain-dependent polymerization to support Wnt-b-catenin signaling. Mol. Cell 63, 498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohieldin AM, Haymour HS, Lo ST, AbouAlaiwi WA, Atkinson KF, Ward CJ, Gao M, Wessely O, and Nauli SM (2015). Protein composition and movements of membrane swellings associated with primary cilia. Cell. Mol. Life Sci 72, 2415–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, and Peng Z (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV (2014). How do cilia organize signalling cascades? Philos. Trans. R. Soc. Lon. B Biol. Sci 369, https://doi.org/10.1098/rstb.2013.0465, 20130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, and Schneider TR (2004). HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr 37, 843–844. [Google Scholar]
- Piazzon N, Maisonneuve C, Guilleret I, Rotman S, and Constam DB (2012). Bicc1 links the regulation of cAMP signaling in polycystic kidneys to microRNA-induced gene silencing. J. Mol. Cell Biol 4, 398–408. [DOI] [PubMed] [Google Scholar]
- Qiao F, and Bowie JU (2005). The many faces of SAM. Sci. STKE 2005, re7. [DOI] [PubMed] [Google Scholar]
- Ramachandran H, Engel C, Müller B, Dengjel J, Walz G, and Yakulov TA (2015). Anks3 alters the sub-cellular localization of the Nek7 kinase. Biochem. Biophys. Res. Commun 464, 901–907. [DOI] [PubMed] [Google Scholar]
- Riccio A, McCauley M, Langelier M-F, and Pascal J (2016). Tankyrase Sterile α Motif domain polymerization is required for its role in Wnt signaling. Structure 24, 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B, Back R, Quinternet M, Bizarro J, Robert MC, Blaud M, Romier C, Manival X, Charpentier B, Bertrand E, et al. (2014). Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res. 42, 2015–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B, Leal-Esteban L, Bernet F, Urfer S, Doerr N, Weimbs T, Iwaszkiewicz J, and Constam DB (2015). Bicc1 polymerization regulates the localization and silencing of bound mRNA. Mol. Cell. Biol 35, 3339–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Yakulov TA, Hashem A, Walz G, and Alkuraya FS (2016). ANKS3 is mutated in a family with autosomal recessive laterality defect. Hum. Genet 135, 1233–1239. [DOI] [PubMed] [Google Scholar]
- Shillingford JM, Leamon CP, Vlahov IR, and Weimbs T (2012). Folate-conjugated rapamycin slows progression of polycystic kidney disease. J. Am. Soc. Nephrol 23, 1674–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagner EE, Bouvrette DJ, Cheng J, and Bryda EC (2009). The polycystic kidney disease-related proteins Bicc1 and SamCystin interact. Biochem. Biophys. Res. Commun 383, 16–21. [DOI] [PubMed] [Google Scholar]
- Taskiran EZ, Korkmaz E, Gucer S, Kosukcu C, Kaymaz F, Koyunlar C, Bryda EC, Chaki M, Lu D, Vadnagara K, et al. (2014). Mutations in ANKS6 cause a nephronophthisis-like phenotype with ESRD. J. Am. Soc. Nephrol 25, 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, and Clardy J (1993). Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Walz G (2017). Role of primary cilia in non-dividing and post-mitotic cells. Cell Tissue Res. 369, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP 4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakulov TA, Yasunaga T, Ramachandran H, Engel C, Müller B, Hoff S, Dengjel J, Lienkamp SS, and Walz G (2015). Anks3 interacts with nephronophthisis proteins and is required for normal renal development. Kidney Int. 87, 1191–1200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.