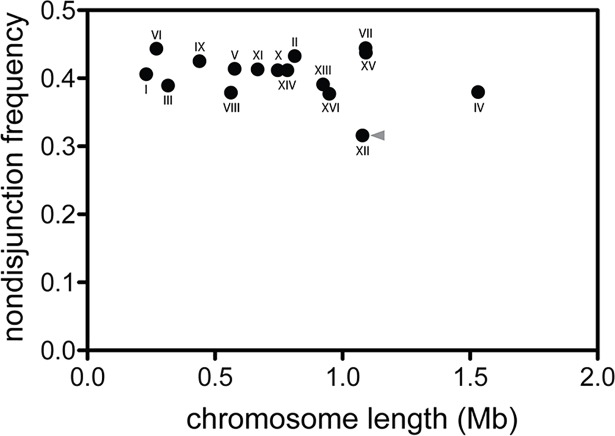
Fig 2. An interspecific cross between S. cerevisiae and S. paradoxus exhibits extremely high rates of meiosis I nondisjunction.
Black circles represent the nondisjunction rate for each of 16 homologous chromosome pairs. Underlying data can be found in S1 Data. Random segregation would correspond to a nondisjunction frequency of 0.5, while perfect segregation would correspond to a nondisjunction frequency of 0.0. Chromosome lengths represent the total amount of alignable sequence per chromosome pair between the genomes of the two species. Length estimates were obtained using the complete genomes of S. cerevisiae strain SK1 (instead of Y55) and S. paradoxus strain CBS432 (instead of N17) because these are the most closely related strains for which end-to-end assemblies are available [22]. The alignable regions of these two genomes are 87.8% identical (9,784,711 out of 11,146,833 sites). The length of Chr XII (indicated by the grey arrowhead) omits the tandem rDNA repeats, which could not be assembled; the true size of Chr XII may be twice as long as presented. Chr, chromosome; rDNA, ribosomal DNA.