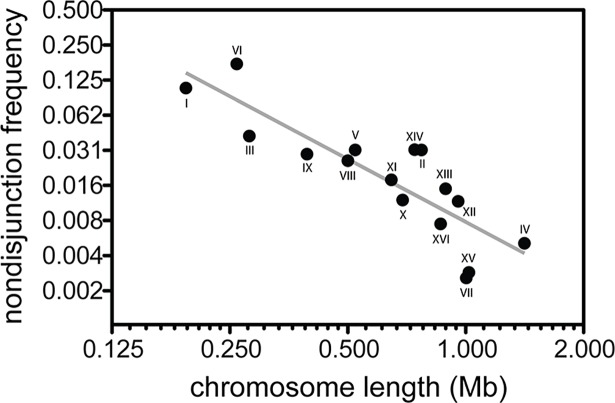
Fig 5. Short chromosomes fail to disjoin at higher rates than long chromosomes in an intraspecific S. paradoxus hybrid.
Circles represent the nondisjunction frequency for each of the 16 homologous chromosome pairs in hybrids formed between the S. paradoxus strains N17 and N44. Underlying data can be found in S1 Data. Chromosome lengths represent the total amount of alignable sequence per chromosome pair between the genomes of each species. Length estimates were obtained using the complete genomes of S. paradoxus strains CBS432 (instead of N17) and N44 [22]. The alignable regions of these two genomes are 98.6% identical (11,358,261 identical sites out of 11,516,349 total sites). The grey line represents the line of best fit: log nondisjunction = −2.12 − 1.79(log length).