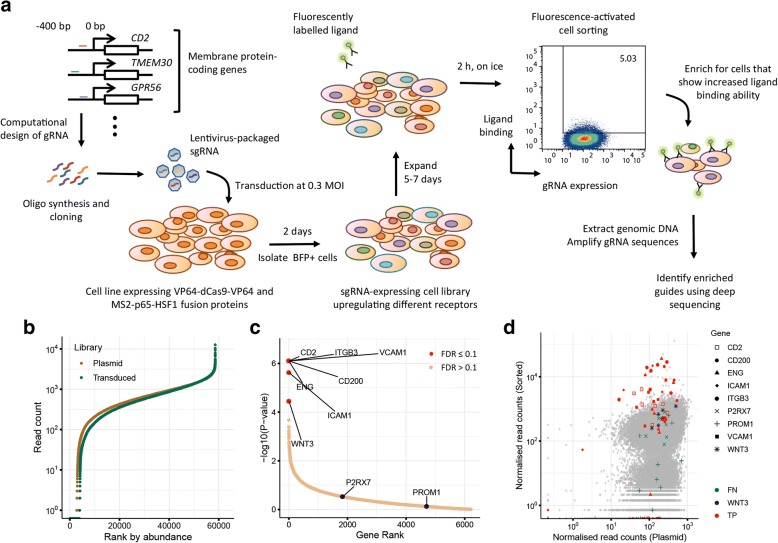
Fig. 2.
Genome-scale enrichment-based extracellular interaction screen identifies multiple antibody targets. a Workflow of CRISPRa screen for identifying extracellular interactions. b Ranked gRNA abundance in the plasmid library and cells transduced with the CRISPRa lentiviral library and cultured for 7 days as determined by raw read counts from deep sequencing of PCR-amplified products. c Plot of transformed P values versus genes in enriched rank order from cells selected using a pool of mAbs targeting eight cell surface proteins: CD2, ITGB3, CD200, VCAM1, ENG, ICAM1, P2RX7, and PROM1. Genes with a false discovery rate (FDR) ≤ 0.1 are indicated with a red dot and labeled. WNT3 was identified as a false positive at this stringency threshold, and P2RX7 and PROM1 as false negatives. d Comparison of gRNA sequencing read counts in fluorescence-sorted cells versus the original plasmid library. gRNA targeting the eight genes and WNT3 are denoted with different shapes, gray dots represent gRNA targeting the promoter regions of all other genes. FN, false negative; FP, false positive; TP, true positive