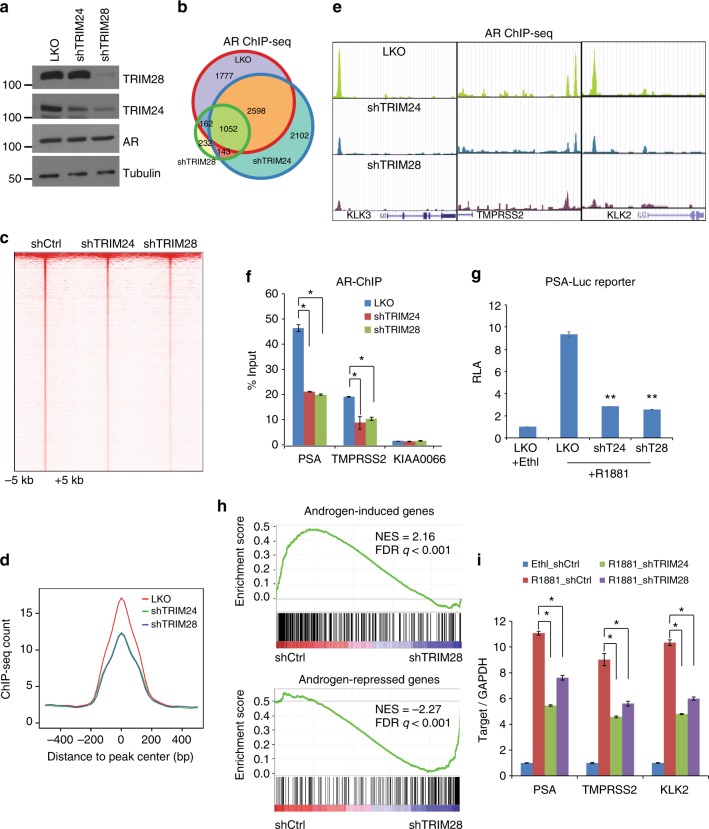
Fig. 5.
TRIM28 enhances AR signaling in prostate cancer. a Western blot analysis of protein lysates of LNCaP cells treated with pLKO, shTRIM24, or shTRIM28 with indicated antibodies. b–e AR ChIP-seq was performed in control, TRIM24-, or TRIM28-knockdown LNCaP cells. Venn diagram (b) shows the overlap of AR binding sites in these cells, where heatmap (c) and intensity plots (d) depict AR ChIP-seq read intensity across all three samples centered (± 5 kb) at AR binding sites found in control LNCaP cells. AR ChIP-seq signals at several known AR-target genes are shown using UCSC Genome Browser Tracks (e). f ChIP-qPCR showing AR enrichment at the PSA and TMPRSS2 enhancers. KIAA0066 was used as negative control. Data shown is mean (± SEM, n = 3). *P < 0.05 by Student’s t-test. g TRIM28 is required for AR transactivation activity. A luciferase reporter construct consisted of PSA enhancer and promoter elements were transfected into pLKO, shTRIM24, and shTRIM28 LNCaP cells. Relative luciferase activities are presented as mean (± SEM, n = 3). **P < 0.01 by Student’s t-test. h TRIM28 positively regulates AR-mediated gene expression program. GSEA was performed to determine the enrichment of AR-induced (upper panel) or -repressed gene set (lower panel) in gene expression dataset profiling control and TRIM28-knockdown LNCaP cells grown in the presence of androgen. i TRIM28 is required for AR-regulated gene expression. LNCaP cells with pLKO, shTRIM24, and shTRIM28 were hormone-starved for 3 days, treated with 1 nM R1881 for 48 h, and subjected to qRT-PCR analysis in comparison to a control LNCaP cells treated with ethanol (Ethl). Data were normalized to GAPDH. Data shown is mean (± SEM, n = 3). *P < 0.05 by Student’s t-test