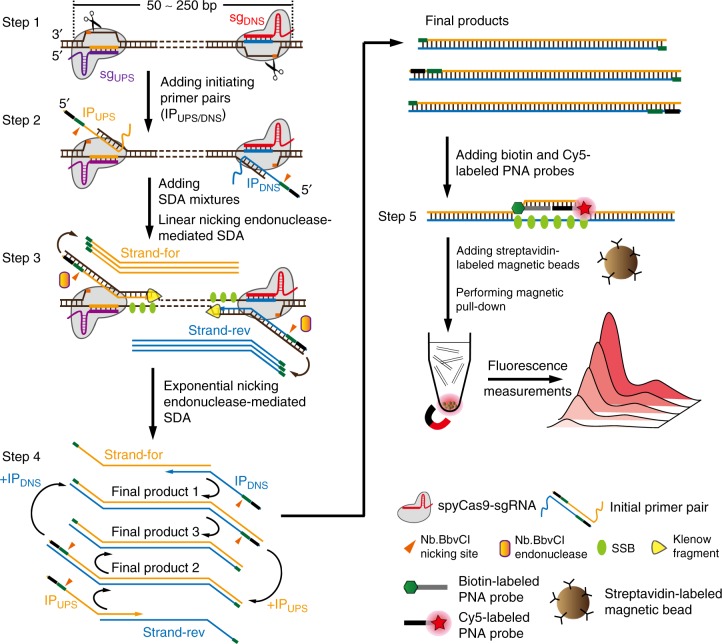
Fig. 1.
Schematic reaction mechanism of CRISDA. Step 1: A pair of Cas9 ribonucleoproteins is programmed to recognize each border of the target DNA and to induce a pair of nicks in both non-target strands. Step 2: A pair of IP primers is introduced and hybridized to the exposed non-target strands. Step 3: After adding SDA mixtures containing KF polymerase (3′– > 5′ exo−), Nb.BbvCI nikase, and single-stranded DNA binding protein TP32 (SSB), linear SDA is initiated from the binding sites of IP primers, giving linearly replaced single strands, the Strand-for and Strand-rev. Step 4: The products, Strand-For and Strand-Rev, are annealed again to the IP primers, which further induce exponential SDA of the selected target sequence. Step 5: The amplicons are quantitatively determined by a PNA invasion-mediated endpoint measurement via magnetic pull-down and fluorescence measurements. The well-characterized S. pyogenes Cas9 with a mutation of HNH catalytic residue (spyCas9H840A nickase) is used as a model. *Two bands will be observed in PAGE analyses, where one corresponds to the final products 1 and 2 with the same length and the other one is product 3