A key virulence factor of influenza A virus is the NS1 protein, which inhibits various cellular processes to facilitate viral gene expression. The NS1 protein is localized in the nucleus and in the cytoplasm during infection. In the nucleus, NS1 has functions related to inhibition of gene expression that involve protein-protein and protein-RNA interactions. While several studies have elucidated the protein interactome of NS1, we still lack a clear and systematic understanding of the NS1-RNA interactome. Here we reveal a nuclear NS1-RNA interactome and show that NS1 primarily binds intronic sequences within a subset of pre-mRNAs, including the RIG-I pre-mRNA that encodes the main cytoplasmic antiviral sensor of influenza virus infection. Our data here further suggest that NS1 is necessary and sufficient to impair intron processing of the RIG-I pre-mRNA. These findings support a posttranscriptional role for NS1 in the inhibition of RIG-I expression.
KEYWORDS: NS1 protein, RNA processing, influenza virus
ABSTRACT
The NS1 protein of influenza A virus is a multifunctional virulence factor that inhibits cellular processes to facilitate viral gene expression. While NS1 is known to interact with RNA and proteins to execute these functions, the cellular RNAs that physically interact with NS1 have not been systematically identified. Here we reveal a NS1 protein-RNA interactome and show that NS1 primarily binds intronic sequences. Among this subset of pre-mRNAs is the RIG-I pre-mRNA, which encodes the main cytoplasmic antiviral sensor of influenza virus infection. This suggested that NS1 interferes with the antiviral response at a posttranscriptional level by virtue of its RNA binding properties. Indeed, we show that NS1 is necessary in the context of viral infection and sufficient upon transfection to decrease the rate of RIG-I intron removal. This NS1 function requires a functional RNA binding domain and is independent of the NS1 interaction with the cleavage and polyadenylation specificity factor CPSF30. NS1 has been previously shown to abrogate RIG-I-mediated antiviral immunity by inhibiting its protein function. Our data further suggest that NS1 also posttranscriptionally alters RIG-I pre-mRNA processing by binding to the RIG-I pre-mRNA.
IMPORTANCE A key virulence factor of influenza A virus is the NS1 protein, which inhibits various cellular processes to facilitate viral gene expression. The NS1 protein is localized in the nucleus and in the cytoplasm during infection. In the nucleus, NS1 has functions related to inhibition of gene expression that involve protein-protein and protein-RNA interactions. While several studies have elucidated the protein interactome of NS1, we still lack a clear and systematic understanding of the NS1-RNA interactome. Here we reveal a nuclear NS1-RNA interactome and show that NS1 primarily binds intronic sequences within a subset of pre-mRNAs, including the RIG-I pre-mRNA that encodes the main cytoplasmic antiviral sensor of influenza virus infection. Our data here further suggest that NS1 is necessary and sufficient to impair intron processing of the RIG-I pre-mRNA. These findings support a posttranscriptional role for NS1 in the inhibition of RIG-I expression.
INTRODUCTION
Nonstructural protein 1 (NS1) of influenza A virus is a major virulence factor that inhibits host antiviral gene expression at several levels. NS1 is synthesized early during influenza virus infection of the host cell, where it accumulates in both the cytoplasm and the nucleus. In the cytoplasm, NS1 prevents signaling through retinoic acid-inducible gene I (RIG-I) (also known as DDX58), which results in inhibition of interferon (IFN) induction (1). NS1 also activates phosphatidylinositol 3-kinase (PI3K) (2, 3) to prevent premature apoptosis during infection (4). Other cytoplasmic functions of NS1 include inhibiting the activation of the antiviral proteins PKR (protein kinase RNA activated) and OAS (2′-5′-oligoadenylate synthetase 1) as well as promoting viral mRNA translation by interacting with translation factors (1). In the nucleus, NS1 binds constituents of the host mRNA processing and nuclear export machineries, leading to inhibition of these processes (5). To perform these tasks, NS1 interacts with the cleavage and polyadenylation specificity factor (CPSF30/CPSF4), the nuclear poly(A) binding protein (PABPN1) (also known as PABII) (6–8), chromatin remodelers such as PAF1 (9), and constituents of the mRNA export machinery, including NXF1 (10). These interactions likely occur in a sequential or coordinated manner as the host mRNAs are being processed and then subsequently exported. While mRNA nuclear export inhibition affects cellular mRNAs, it does not impair nuclear export of viral mRNAs (10). NS1 also inhibits cellular pre-mRNA splicing by interacting with U6atac snRNA (11) but promotes splicing of the viral M mRNA segment and its nuclear export through host nuclear speckles (12). Thus, through multiple processes, NS1 efficiently provides a favorable cellular environment for influenza virus infection.
The multifunctionality of NS1 impairs the immune response to favor virus replication (1). In fact, influenza virus lacking NS1 can properly replicate only in immunocompromised hosts (13). The ability of NS1 to interact with RNA and various cellular proteins is likely due to its structural plasticity, multiple intracellular locations (nucleus and cytoplasm), and posttranslational modifications. NS1 can adopt diverse structures, from homodimers to oligomers (14), with multiple spatial conformations (15), providing enough protein surface diversity for binding multiple proteins and RNA. Several studies have been conducted that elucidated the protein interactome of NS1 (16–21). However, we still lack a clear understanding on the NS1-RNA interactome. To this end, we show that NS1 interacts with specific subsets of cellular mRNAs during infection and that this binding occurs primarily on introns, supporting a cotranscriptional association of NS1 with nascent host transcripts. Among the enriched immune-related transcripts is the RIG-I pre-mRNA, whose processing appears to be inhibited by NS1. Our results suggest that NS1 interacts directly with the RIG-I pre-mRNA to prevent its expression. In combination with previous reports of NS1 inhibition of the RIG-I protein, our results support that NS1 mounts a two-pronged attack on RIG-I, a key factor for interferon expression and immunity.
RESULTS
Genome-wide identification of NS1 target RNAs during influenza virus infection.
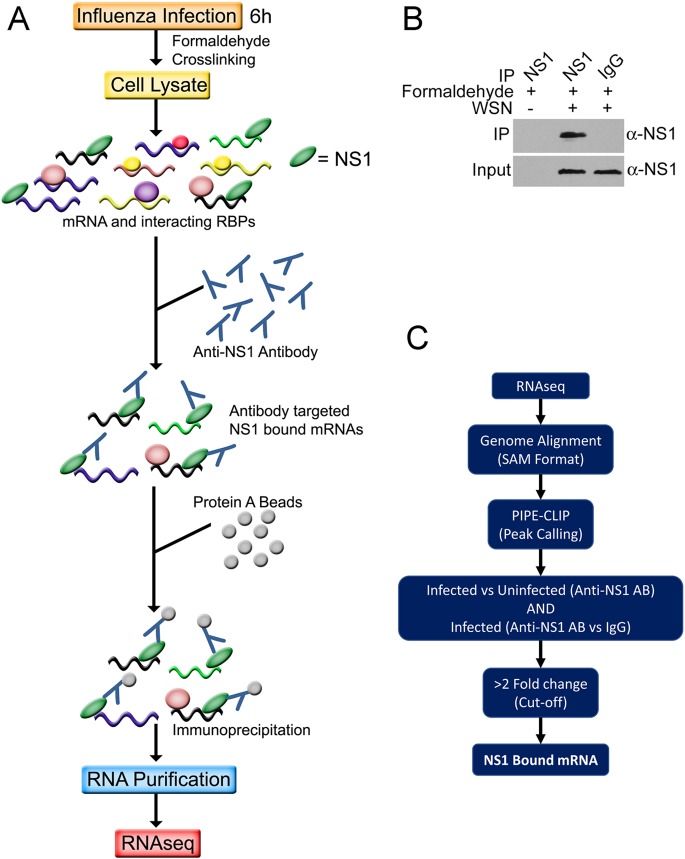
To globally identify the RNA targets of NS1 midway through infection, we employed a formaldehyde crosslinking RNA immunoprecipitation (fRIP) procedure (Fig. 1A). NS1 has been suggested to bind to RNAs not only by virtue of its RNA binding domain but also indirectly through its associations with other proteins (1, 22). Therefore, we first employed formaldehyde crosslinking over UV crosslinking, as the latter requires direct RNA-protein interactions. Six hours following infection of A549 cells with influenza A/WSN/33 (H1N1) (WSN) virus, we added formaldehyde to covalently link RNA-protein complexes. NS1-bound mRNAs were purified with an anti-NS1 antibody (Fig. 1B), and PIPE-CLIP analysis (23) was conducted to identify NS1-associated transcripts (Fig. 1C; see also Table S1 in the supplemental material). We defined the NS1-associated RNAs to be those transcripts that satisfied three criteria: (i) they were >2-fold enriched relative to IgG controls in infected cells, (ii) they were >2-fold enriched relative to anti-NS1 RIP in uninfected cells, and (iii) they passed these thresholds in two independent biological replicates.
FIG 1.
fRIP-seq protocol. (A) Schematic diagram of the fRIP-seq protocol. Details are given in Results and in Materials and Methods. (B) Immunoprecipitation followed by Western blot validation of the anti-NS1 antibody immunoprecipitations of NS1 under fRIP conditions. (C) Pipeline for bioinformatic analysis. See Materials and Methods for details. AB, antibody; RBPs, RNA-binding proteins.
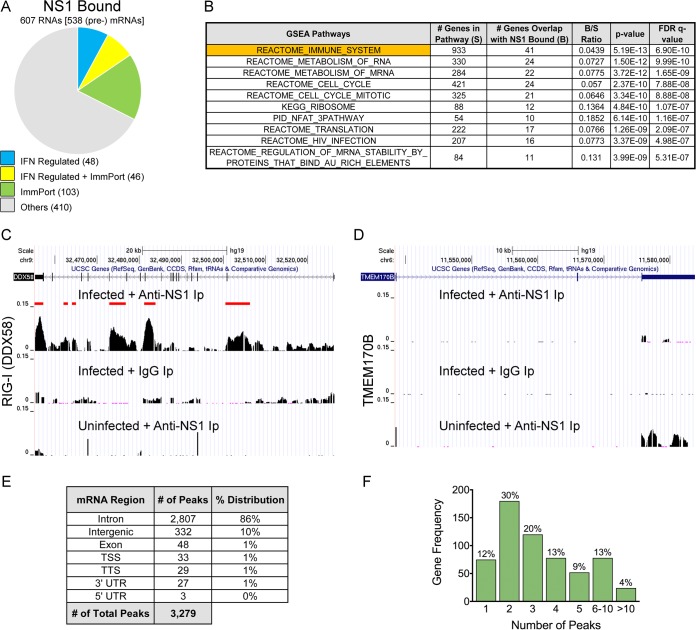
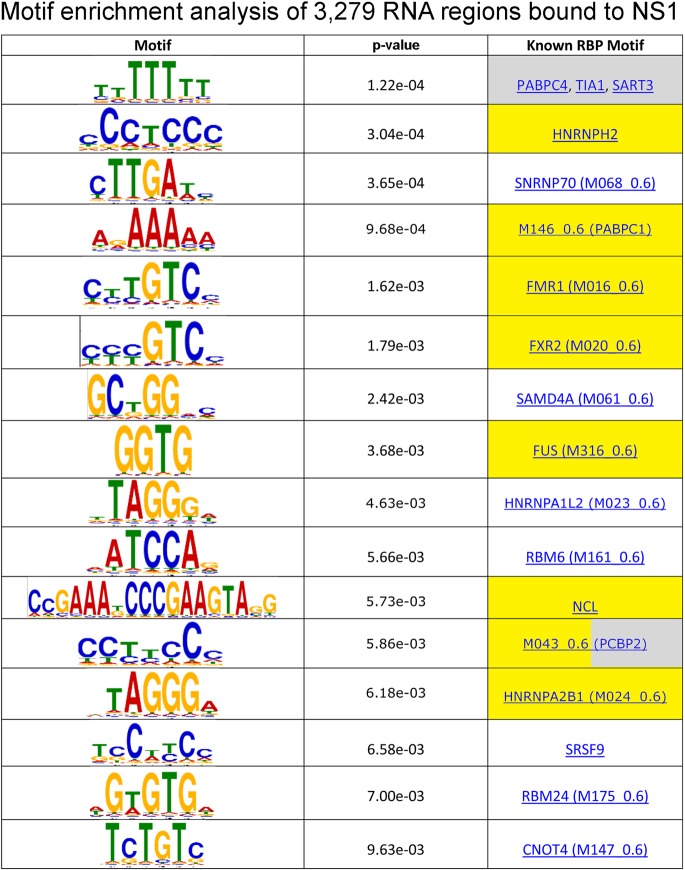
Using these cutoffs, we identified 607 NS1-bound RNAs (Fig. 2A), among which are 538 (pre-)mRNAs. Only 3% of the upregulated RNAs at 6 h postinfection (hpi), compared to uninfected cells, were NS1 bound, as measured by microarray analysis (Table S2). The upregulated NS1-bound RNAs represented 8% of the 538 NS1-bound mRNAs. Therefore, NS1 binding does not correlate with mRNAs whose steady-state levels are increased at 6 h postinfection. Gene set enrichment analysis (GSEA) showed an enrichment of immune-related transcripts in the NS1-bound data set, including RIG-I (197/607 genes) (Fig. 2A and B and Table S1). We observed multiple enriched peaks across the RIG-I RNA (Fig. 2C, red bars) but not over negative-control samples (Fig. 2D, Infected + IgG Ip and Uninfected + Anti-NS1 Ip) or negative-control genes (e.g., TMEM170B) (Fig. 2D). Interestingly, peaks were primarily in the introns, and this intronic pattern of NS1 binding was not unique to RIG-I. In fact, ∼86% of the NS1 binding sites were observed in introns (2,807/3,279 peaks) (Fig. 2E). This observation suggests that NS1 interacts with pre-mRNAs, consistent with previous reports showing NS1 effects on splicing (11, 24, 25). Given that most splicing occurs cotranscriptionally (26), these data further suggest that NS1 interacts with nascent transcripts. Moreover, the lower abundance of introns than of exons in total RNA supports that the identified RNAs result from bona fide interactions and are not simply due to artifactual overrepresentation of abundant RNAs. The number of peaks identified was considerably higher than the number of genes (3,279 compared to 607), indicating that most genes had multiple NS1 peaks. In fact, only ∼12% of the genes that were identified contained a single NS1 peak, and nearly 40% had four or more peaks (Fig. 2F). We further searched for enriched motifs in the NS1-bound regions, but no clear consensus sequence was identified, although the identified regions contain predicted sites for specific RNPs, some of which are regulated by IFN and/or involved in immunity (Fig. 3). Taken together, these data suggest that NS1 binds cotranscriptionally to specific host pre-mRNAs during influenza virus infection.
FIG 2.
NS1 associates with introns of pre-mRNAs. (A) Pie chart showing the distribution of NS1-associated RNAs that are classified as interferon (IFN) regulated or as immune related by the Interferome and ImmPort databases. (B) GSEA enrichment categories for NS1-associated RNAs. (C) Sequence traces of peaks across the RIG-I (DDX58) gene. The middle and bottom traces are the infected IgG RIP and uninfected anti-NS1 RIP negative controls. (D) Traces as in panel C except for the expressed but noninteracting RNA TMEM170B. (E) Gene segment annotations for the identified NS1-associated RNA peaks. TSS, transcription start site. TTS, transcription termination site. (F) Number (and percentage) of peaks representing NS1 binding sites per RNA identified.
FIG 3.
Motif analysis of NS1-immunoprecipitated RNAs. NS1 interaction peaks were assessed for enriched motifs using MEME-SUITE. Known RNA binding proteins that bind similar motifs are also listed (CISBP-RNA database). A subset of these RNA binding proteins is either IFN regulated (yellow) or listed in the ImmPort database (gray).
NS1 directly binds to RNA targets.
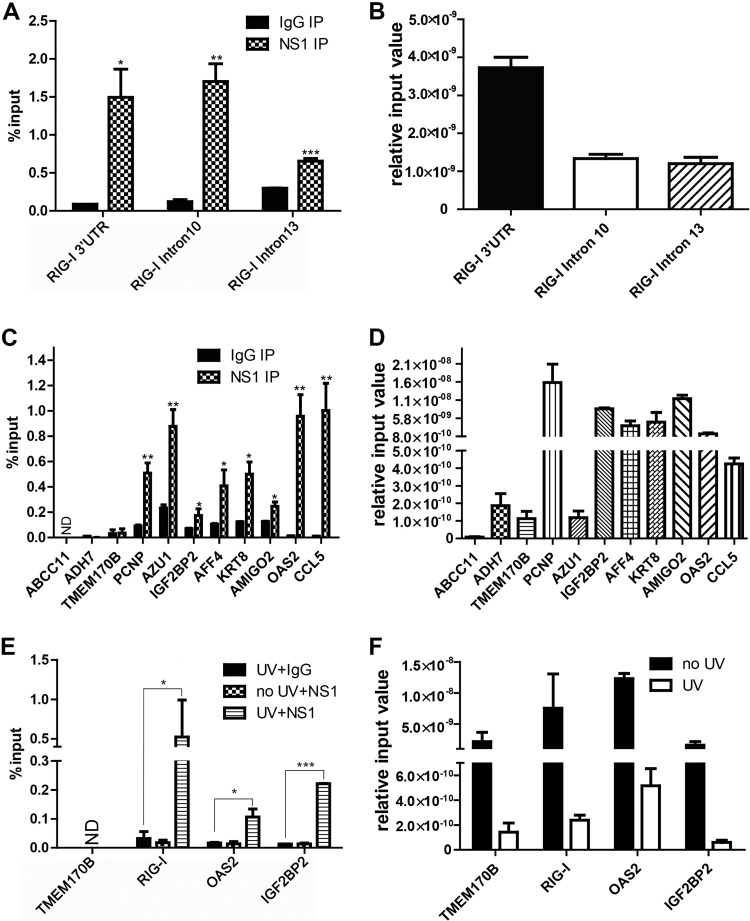
We next validated the specificity of NS1 binding to RIG-I and of additional transcripts using fRIP and reverse transcription-quantitative PCR (RT-qPCR). Consistent with the global analyses, we observed binding over the background for the RIG-I 3′ untranslated region (UTR) and intron 10 but to a lesser extent to intron 13 (Fig. 4A and B). In addition, association with a number of additional RNAs was verified, while negative-control RNAs not identified in our global analysis (ABCC11, ADH7, and TMEM170B) were not immunoprecipitated (Fig. 4C and D). This corroborates the association of NS1 with specific cellular RNAs observed in our fRIP-seq (fRIP and transcriptome sequencing [RNA-seq]) experiments. Next, we used a UV-crosslinking scheme to test potential direct interactions between NS1 and some of its RNA targets. Unlike formaldehyde crosslinking, UV will crosslink only direct RNA-protein interactions. Moreover, because live cells are exposed to UV, the observation of a UV-dependent interaction demonstrates that the RNA-protein interaction occurs in living cells and does not result from binding in extracts after lysis (27). Indeed, we observed enrichment of RIG-I, OAS2, and IGF2BP2 RNAs over IgG and uncrosslinked controls (Fig. 4E and F). Therefore, we conclude that NS1 binds directly to these transcripts in influenza virus-infected cells.
FIG 4.
Validation and direct binding of NS1 to RNAs. (A) Formaldehyde RIP and RT-qPCR were performed with NS1 or IgG control antibody. Because the RNA is sheared prior to RIP, the indicated regions of RIG-I can be independently assessed. Data are mean values ± standard deviations (SD) (n = 3). (B) Bar graph showing relative input levels for the RIG-I 3ʹ UTR and RIG-I introns 10 and 13 in panel A. (C) Validation by RT-qPCR of the indicated targets identified by fRIP-seq. ABCC11, TMEM170B, and ADH7 were not identified in our fRIP-seq analyses and serve as negative controls. Data are mean values ± SD (n = 3). (D) Bar graph showing relative input levels for the various RNAs depicted in panel C. (E) UV-crosslinking immunoprecipitation results for the indicated genes. TMEM170B is a negative-control transcript, and no-UV and IgG immunoprecipitations are negative-control samples. (F) Bar graphs showing relative input levels for the RNAs depicted in panel E. All lysates for which RIP was performed with NS1 or IgG antibody were derived from cells infected with influenza virus (WSN). Data are mean values ± SD (n = 3) (ND, not detected). In all panels, data were analyzed for statistically significant differences using unpaired, two-tailed Student’s t test, and significance is annotated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Regulatory impact of NS1 on its mRNA targets.
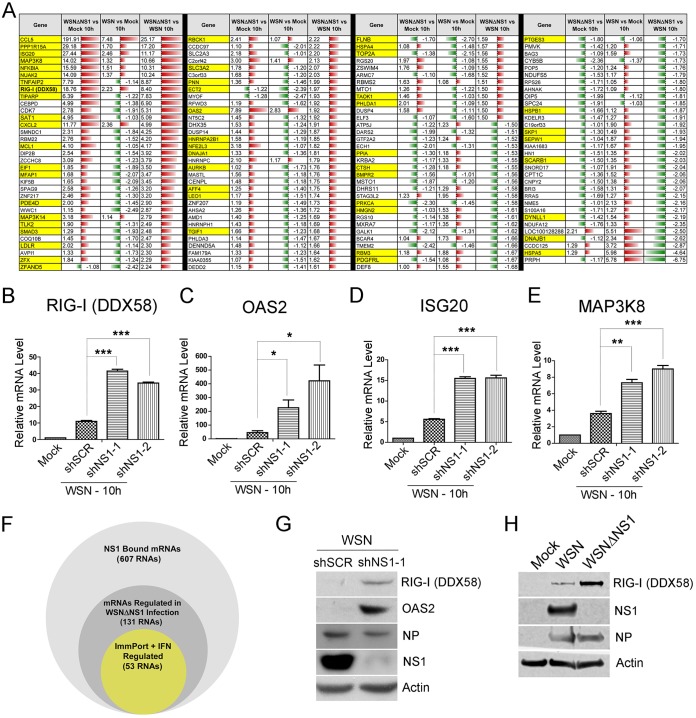
To assess the effects of NS1 on the levels of its bound RNAs, we compared expression profiles of A549 cells infected for 10 h with wild-type and NS1-deleted mutant WSN (WSNΔNS1) viruses. Among NS1-bound RNAs, 22% (131/607) showed differential expression between WSN and WSNΔNS1 infection (Fig. 5A and F; see also Table S3 in the supplemental material). Moreover, 40% (53/131) of these differentially expressed genes are immune or interferon regulated (Fig. 5A and F, yellow). Notably, among NS1-bound RNAs that were differentially expressed, the majority (∼60%) was upregulated upon NS1 deletion. Among the immune/interferon-regulated genes, 72% (38/53) were upregulated, in agreement with previous findings and with the known function of NS1 as a suppressor of immune-related genes (1).
FIG 5.
Expression changes in NS1-bound RNAs. (A) Table showing changes in expression of the genes bound by NS1. Microarray profiling was performed at 10 hpi for the WSN and WSNΔNS1 strains (see Table S3 in the supplemental material). The table in this panel depicts changes relative to mock-infected or infected cells, as indicated. Transcripts highlighted in yellow are found in the Interferome and/or ImmPort database. (B to E) Independent validation of the NS1-dependent changes in gene expression performed by WSN infection of A549 cells stably expressing one of two NS1-targeting shRNAs or a nontargeting scrambled (SCR) control followed by RT-qPCR to quantify levels of the depicted mRNAs. (F) Venn diagram showing the portions of NS1-bound RNAs (light gray) that are differentially regulated during WSNΔNS1 infection compared to WSN infection (dark gray) and those that were identified in the Interferome and ImmPort databases (yellow). (G and H) Western blots confirming that changes in RNA levels of OAS2 and RIG-I are also observed at the protein level. (G) WSN-infected A549 cells stably expressing NS1-targeting shRNA or nontargeting scrambled shRNA were used. (H) Cells were infected with WSN or WSNΔNS1 as described above for panel A (n = 3) (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
We have also employed a complementary approach to confirm the effect of NS1 on the expression of several important immune regulators: RIG-I, OAS2, ISG20, and MAP3K8. We generated A549 cells that constitutively express one of two short hairpin RNAs (shRNAs) against NS1 (shNS1-1 or shNS1-2) or a scrambled control (shSCR) and infected them with WSN (Fig. 5B to E). As expected, we observed enhanced expression of these antiviral genes upon viral infection (comparing mock infection to shSCR). NS1 knockdown with either NS1 shRNA led to elevated levels of each of these transcripts compared to the scrambled controls. We then examined the protein levels of two of these genes, OAS2 and RIG-I, and observed similar effects (Fig. 5G and H). With the results presented above, these observations are consistent with NS1 binding to antiviral pre-mRNAs resulting in inhibition of expression. However, as NS1 is also able to inhibit transcriptional activation of innate immune genes (1, 28), the relative contributions of NS1 to transcriptional versus posttranscriptional inhibition of host protein levels are not clear.
NS1 alters RIG-I processing.
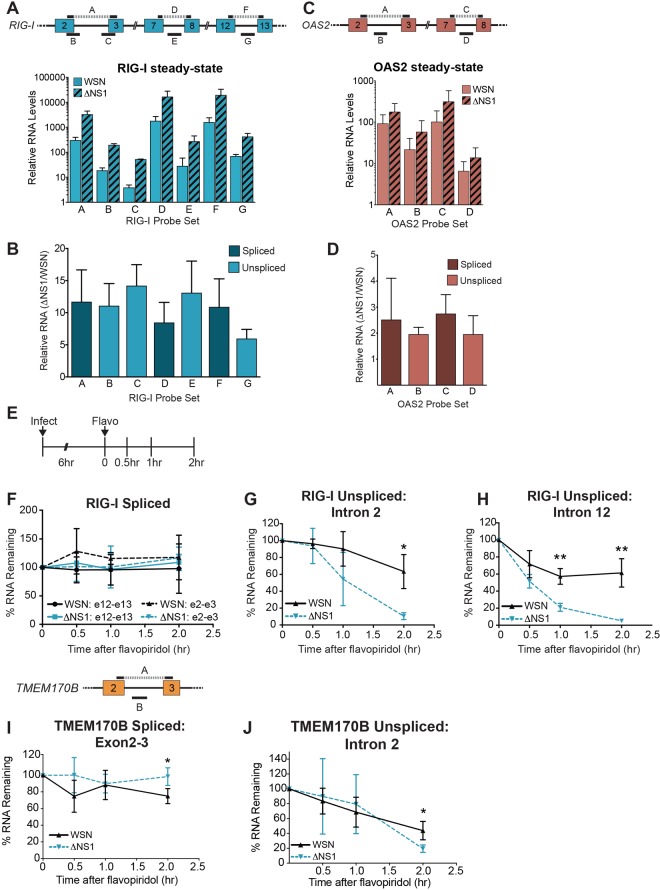
We next focused on the functional consequences of NS1 binding to the RIG-I pre-mRNA. We chose to focus on RIG-I as a representative NS1 target because of its central importance to immunity during influenza virus infection. Given that NS1 binds introns, we hypothesized that NS1 cotranscriptionally associates with RNAs to affect the earliest stages of RNA biosynthesis. Initially, we examined steady-state RIG-I RNA levels using seven RT-PCR primer sets specific for spliced and unspliced RIG-I RNAs (Fig. 6A, top). Comparison of the steady-state levels of these RNAs in WSNΔNS1-infected cells to those in WSN-infected cells (6 hpi) showed that levels of both the spliced and unspliced RNAs increased from ∼6- to 13-fold in the WSNΔNS1 strains (Fig. 6A and B). Similar results were observed for OAS2, although the magnitude of the increase was only ∼2-fold (Fig. 6C and D). The similarities in upregulations of both spliced and intron-containing RNAs strongly support previous reports that NS1 blunts the activation of immune signaling pathways. That is, in the WSN strain, transcriptional activation of the RIG-I and OAS2 genes is repressed, whereas the WSNΔNS1 strain loses this innate immune inactivation (1, 28).
FIG 6.
NS1 deletion alters RIG-I intron processing in infected cells. (A, top) Gene diagram and placement of RT-qPCR amplicons across the RIG-I gene. (Bottom) Steady-state analysis of RIG-I spliced and intronic regions at 6 hpi. All values were first normalized to the value for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are means ± SD (n = 3). (B) The ΔNS1 steady-state values from panel A are presented relative to the WSN values, which were set to 1 for each amplicon in each replicate. Data are means ± SD (n = 3). (C) Same as panel A except that OAS2 was examined. (D) Same as panel B except that OAS2 was examined. (E) Time course of flavopiridol experiments. (F) Decay curves for RIG-I spliced exons 2 and 3 (amplicon A from panel A) and spliced exons 12 and 13 (amplicon F) in WSN or WSNΔNS1 infection, as indicated. (G) Rates of disappearance of RIG-I intron 2 (amplicon C) in WSN or WSNΔNS1 infection, as indicated. (H) Same as panel G except that intron 12 (amplicon G) was assessed. (I, top) Gene diagram and placement of RT-qPCR amplicons across the TMEM170B gene, as a negative control. (Bottom) Rates of disappearance of spliced TMEM170B (amplicon A) in the WSN or WSNΔNS1 strain. (J) Rates of disappearance of TMEM170B intron 2 (amplicon B) in the WSN or WSNΔNS1 strain. In panels F through J, all time points were analyzed for statistically significant differences using unpaired, two-tailed Student’s t test, and significance is annotated (*, P < 0.05; **, P < 0.01).
While transcription of innate immune genes is blunted in the presence of NS1, some transcripts are still synthesized, as evidenced by the association of NS1 with intronic sequences in RIG-I pre-mRNA (Fig. 2C). Assessment of our hypothesized posttranscriptional effects of NS1 on RNA processing are confounded by the higher steady-state levels of mRNA than of pre-mRNA and by the differences in transcription in the presence and absence of NS1 (Fig. 6A to D). To experimentally separate these activities, we performed a transcription inhibition assay to assess the rates of disappearance of RIG-I introns and spliced exons. Six hours after infection with WSN or WSNΔNS1, we added flavopiridol, an RNA polymerase II transcription inhibitor, and we monitored RIG-I transcript levels over time (Fig. 6E). As expected, we observed no statistically significant differences in the rates of decay of the spliced RIG-I mRNAs (Fig. 6F). In contrast, we observed a higher rate of disappearance of the unspliced introns in the WSNΔNS1 strain than in the WSN strain (Fig. 6G and H). We emphasize that these introns of RIG-I are present at higher levels in the WSNΔNS1 strain (Fig. 6A and B), yet they disappear more quickly. In contrast, we observed no such differences with TMEM170B (Fig. 6I and J), which does not associate with NS1 (Fig. 2D and Fig. 4C and E).
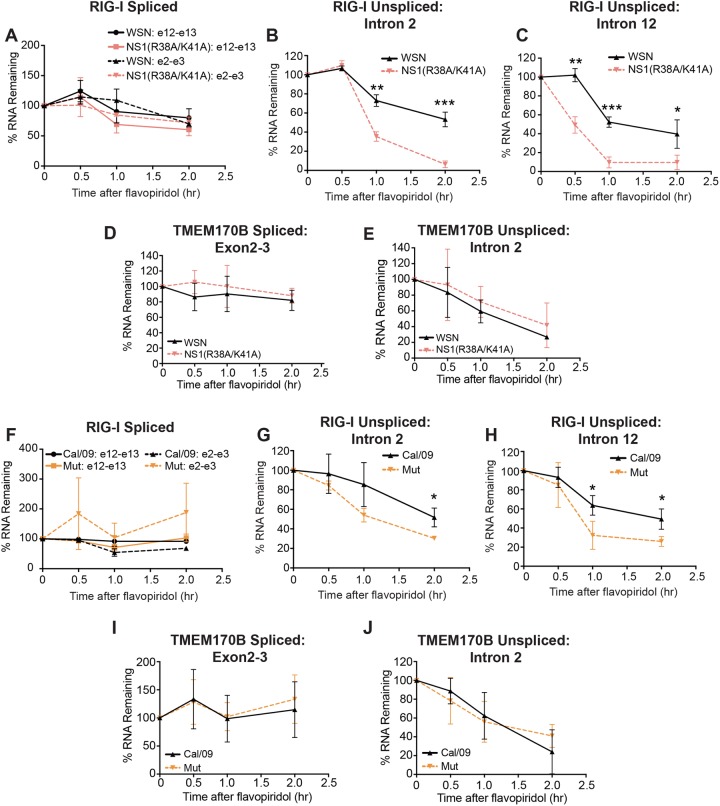
NS1 deletion will have far-reaching effects on the virus, so we next performed the flavopiridol experiments on a mutant strain containing two point mutations in NS1 (R38A/K41A) that abrogate its ability to bind RNA (29). The RNA binding mutant mirrored NS1 deletion on RIG-I intron disappearance rates (Fig. 7A to C) and, as expected, had no effect on TMEM170B (Fig. 7D and E). Thus, RNA binding is necessary for the apparent effects of NS1 on RIG-I intron processing.
FIG 7.
NS1 RNA binding is necessary for its effects on RIG-I processing, but CPSF30 interaction is not. (A to E) WSN and NS1 RNA binding mutant (R38A/K41A) strains were compared in a flavopiridol transcription shutoff analysis. See Fig. 6E to J for details. Data are means ± SD (n = 3). (F to J) Same as for panels A to E except that we used the Cal/09 strain (black lines) and compared it to Cal/09 encoding the NS1 mutant (R108K/E125D/G189D) (“Mut,” orange lines) that restores NS1 binding to CPSF30. In all panels, time points were analyzed for statistically significant differences using unpaired, two-tailed Student’s t test, and significance is annotated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
NS1 inhibits 3′-end formation (6–8), which is linked to splicing. Therefore, it is reasonable to propose that the effects of NS1 on splicing result from its inhibition of 3′-end formation. Therefore, we tested the influenza A/California/2009 (A/Cal/09) virus strain, which expresses NS1 that lacks the ability to bind CPSF30 and to inhibit polyadenylation (30). We compared this to an influenza A/Cal/09 virus mutant strain that expresses an NS1 triple mutant protein (R108K/E125D/G189D) that restores CPSF30 binding (30). Similar to WSN, wild-type Cal/09 displayed a delayed disappearance of introns, and restoration of CPSF30 binding did not further decrease the rate of intron loss (Fig. 7F to H). In fact, restoration of CPSF30 binding showed a slightly faster intron disappearance, which may explain, at least in part, the slight attenuation of this mutant virus with restored CPSF30 inhibition in vivo (30). Once again, none of these NS1 variants affected the negative-control pre-mRNA TMEM170B (Fig. 7I and J). These results strongly suggest that the effect of NS1 on RIG-I intron disappearance is independent of its ability to bind CPSF30.
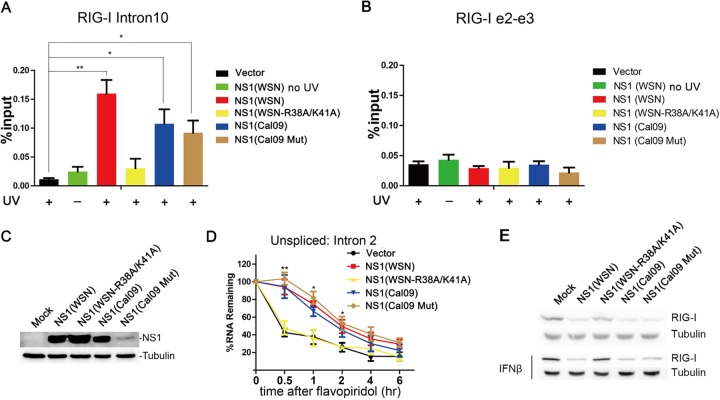
To determine whether NS1 is sufficient to bind and inhibit intron processing of RIG-I mRNA, we transfected NS1 from WSN into HEK293T cells and performed UV crosslinking followed by RIP. Similar to infected cells, we show that NS1 alone binds to an intronic region of RIG-I pre-mRNA (Fig. 8A), as opposed to exons (Fig. 8B). In contrast, when an RNA binding-defective NS1 protein was expressed (NS1 R38A/K41A) (29), binding to the RIG-I intron was lost. In addition, we expressed NS1 from the Cal09 strain, which does not interact with CPSF30, and an NS1 mutant (R108K/E125D/G189D) that is known to restore CPSF30 binding (30). This NS1 mutant is expressed at low levels compared to wild-type Cal09 NS1 (Fig. 8C) (30). However, we found that both wild-type Cal09 NS1 and the mutant that restored CPSF30 binding interact with the RIG-I intronic region. Thus, we show that the RNA binding activity of NS1 is required for NS1 binding to the RIG-I intronic region, while CPSF30 does not appear to be necessary for this interaction. Low levels of NS1 expression, as shown in the case of the mutant Cal 09 NS1 protein that acquired the CPSF30 binding compared to the other NS1s expressed in parallel, was sufficient to bind the RIG-I intronic region (Fig. 8A and C).
FIG 8.
NS1 is sufficient to inhibit RIG-I pre-mRNA processing. (A and B) UV-crosslinking immunoprecipitation with anti-NS1 antibody was performed in HEK293T cells transfected with the vector or plasmids encoding the various depicted NS1 proteins. Vector and non-UV-treated NS1 (WSN) RIPs are negative controls. RIG-I intron 10 or spliced exons 2 and 3 were detected by RT-qPCR. Data are mean values ± SD (n = 3). (C) Western blot of HEK293T cell lysates expressing the various depicted NS1 proteins. (D) Rates of disappearance of RIG-I intron 2 in HEK293T cells transfected with plasmids encoding the various depicted NS1 proteins after flavopiridol treatment. Intron 2 was detected by RT-qPCR as described in the legend of Fig. 6. Data are mean values ± SD (n = 3). Data were analyzed for statistically significant differences using unpaired, two-tailed Student’s t test, and significance is annotated (*, P < 0.05; **, P < 0.01). (E) Western blots performed as described above for panel C except that IFN-β was added, as indicated, and RIG-I was detected. Blots containing samples not treated with IFN were overexposed, compared to IFN-treated cells, to allow clear detection of RIG-I protein levels.
We then performed a transcription inhibition assay, as shown in Fig. 6, to assess the rates of disappearance of RIG-I intron 2 in the presence of NS1 proteins from the different strains (Fig. 8D). Wild-type NS1 from the WSN or Cal/09 strain and the Cal/09 mutant, which restored CPSF30 binding, showed similar disappearance rates (Fig. 8D). On the other hand, the RNA binding-defective NS1 protein showed accelerated intron disappearance, as in control cells (Fig. 8D). These results are similar to those for cells infected with the corresponding viruses and support the conclusion that NS1 proteins from different strains have the ability to prevent RIG-I pre-mRNA processing. In addition, these observed NS1 effects at the pre-mRNA level reflected similarly on the levels of RIG-I protein in either the absence or presence of IFN-β, which mimics infection conditions (Fig. 8E). Our results therefore suggest that in addition to affecting the RIG-I pathway by blocking its signaling function (1), NS1 inhibits the maturation of RIG-I pre-mRNA in the nucleus.
DISCUSSION
RNA viruses, such as influenza viruses, enter the cell’s cytoplasm, where their genomic RNA is recognized by RIG-I, triggering its activation (31–33). RIG-I is K63-linked polyubiquitinated by TRIM25 (34) and RIPLET (35) and can also bind free K63 polyubiquitin. Subsequently, RIG-I interacts with the mitochondrial antiviral signaling protein (MAVS) and then activates NF-κB and IRF3, resulting in transcription activation of antiviral genes, among which is interferon (36, 37). This process is disrupted by NS1, which binds RIG-I and TRIM25 to inhibit signaling and interferon expression (34, 38). Recently, studies using bimolecular fluorescence complementation and superresolution microscopy indicated that NS1 forms a complex with RIG-I and TRIM25, which likely prevents the formation of the RIG-MAVS complex and signaling (39). Thus, NS1 inhibition of the RIG-I pathway is critical for impairing the innate immune response, and it provides an optimal environment for influenza virus replication.
We propose that in addition to its cytoplasmic role in the RIG-I pathway, NS1 acts in the nucleus to block RIG-I expression. NS1 binds the RIG-I pre-mRNA at intronic regions and at the 3ʹ UTR, and its presence decreases the rate of RIG-I intron loss. While the inhibitory function of RIG-I signaling by cytoplasmic NS1 results in transcription downregulation of immune-related genes, this inhibition is not complete. Therefore, the pre-mRNAs that are still synthesized can be bound by NS1, and their processing is inhibited. Consequently, RIG-I mRNA and protein expressions are dampened, which would further prevent interferon expression. These mechanisms point to a remarkable versatility and efficiency of the NS1 protein, which functions in the nucleus and in the cytoplasm to robustly inhibit interferon production and immunity by targeting both RIG-I protein function and RIG-I mRNA expression.
NS1 targets not only the RIG-I mRNA but also a subset of mRNAs that encode immune and/or antiviral factors. Most of the binding sites occurred in intronic sequences, and multiple binding sites were observed in a single pre-mRNA. At present, we do not know how specificity for binding to some mRNAs and not to others is achieved by NS1. Interestingly, among the pre-mRNAs bound by NS1 is OAS2, which is one of the OAS proteins that are 2′,5ʹ-oligoadenylate (2-5A) synthases of the (OAS-)RNase L antiviral system and are interferon induced (40). OAS proteins are pattern recognition receptors for viral double-stranded RNA (dsRNA). This interaction activates OAS, which synthesizes 2-5A, which in turn activates RNase L to cleave viral and cellular single-stranded RNAs (ssRNAs). These effects result in inhibition of virus replication. The mRNA from the RNase ISG20, which targets single-stranded RNA, also interacts with NS1. ISG20 is induced by interferon, and its antiviral function is related to its exonuclease activity (41). Another mRNA bound by NS1 encodes the chemokine CCL5 (RANTES), which recruits leukocytes to sites of inflammation and maintains CD8 T cell responses during chronic viral infection (42). CXCL2 is another chemokine whose mRNA is targeted by NS1, and it appears to have a role in the inflammatory response during virus infection (43). Other immune-related and interferon-regulated pre-mRNAs are also bound and downregulated by NS1, suggesting that this subset of pre-mRNAs is suppressed at the processing level to ensure a powerful inhibition of immunity.
MATERIALS AND METHODS
Cell lines, viruses, reagents, plasmids, and antibodies.
A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Where indicated, flavopiridol was added to the medium at 1 μM. HEK293T cells (ATCC) and IFN (catalog number 10704-HNAS; Sino Biological) were used for Fig. 8. Influenza virus A/WSN/33 (H1N1) strains were used. Influenza virus WSNΔNS1 was generated as previously described (44). WSN(NS1 R38A/K41A) was generated as reported previously (29). The Cal/09 wild-type strain and the Cal09(NS1 R108K/E125D/G189D) mutant strain were generated as previously described (30). The corresponding NS1 proteins from these viruses were cloned into the pCAGGS vector as reported previously (29, 30). The following antibodies were used for immunoblot analysis: DDX58 or RIG-I (catalog number 20566-1-AP; Proteintech), OAS2 (catalog number 19279-1-AP; Proteintech), actin (Sigma), and NS1 (a gift from J. A. Richt, National Animal Disease Center, IA) antibodies.
Transfection and Western blotting.
As shown in Fig. 8, HEK293T cells were transfected with the indicated plasmids using polyetherimide (Sigma-Aldrich). After 24 h, cells were treated with IFN-β (500 U/ml) for 6 h, and cell lysates were subjected to Western blotting, as previously described (45).
Formaldehyde RNA immunoprecipitation and RNA-seq.
Transcripts associated with NS1 were isolated as described previously (46). Briefly, A549 cells were infected with influenza virus A/WSN/33 at a multiplicity of infection (MOI) of 5. After 6 h of infection, cells were treated with 0.5% formaldehyde for 10 min at room temperature and quenched with 2.5 M glycine (pH 7.0) for 5 min at room temperature. Cells were lysed with RIP assay (RIPA) buffer (1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM sodium chloride, 50 mM Tris-Cl [pH 8.0], 2 mM EDTA) and sonicated. A total of 1% was saved (input), and immunoprecipitation was performed with anti-NS1 antibody or rabbit IgG overnight. After five washes (twice with RIPA buffer, twice with RIPA buffer–1 M urea, and once with RIPA buffer), crosslinks were reversed with reverse buffer (10 mM Tris-HCl [pH 6.8], 5 mM EDTA, 10 mM dithiothreitol [DTT], 1.0% SDS), and samples were treated with proteinase K at 42°C for 1 h and at 65°C for 1.5 h. RNA was harvested in TRIzol (Invitrogen), transcribed into cDNA, and analyzed by qPCR. The recovery efficiency (percentage of the input) of the RNA-IP was calculated as 2CT sample − CT input, and the ratio of the recovery efficiency of NS1-IP to that of IgG-IP is shown as fold enrichment.
WSN infection and microarray.
A549 cells were infected with A/WSN/1933 at an MOI of 5 for 6 h or 10 h. Total RNA was isolated with TRIzol and probed for gene expression using Illumina Human BeadChip 22k according to the manufacturer’s instructions.
Establishment of shRNA-expressing stable cell lines.
shRNA-expressing plasmids were constructed in the pLKO vector (47). Lentiviruses expressing shRNA were produced by polyethylenimine (PEI)-mediated, three-plasmid transfection of HEK293T cells. Briefly, HEK293T cells (2.4 × 107 cells) were transfected with 12 μg pLKO-shRNA, 7.5 μg pspAX2, and 4.5 μg pMD2.0G and cultured for 48 h. Viral supernatants were harvested and filtered through a 0.45-μm filter. A549 cells were transduced by lentivirus in the presence of 8 μg/ml Polybrene; 1 μg/ml puromycin was added at 24 h posttransduction. shRNA sequences were GTTCGAGTCTCTGAAACTCTA (shNS1-1), GTCTCTGAAACTCTACAGAGA (shNS1-2), and CAACAAGATGAAGAGCACCAA (shSCR).
Quantitative PCR.
Total RNA was isolated from A549 cells with TRIzol (Invitrogen) and reverse transcribed into cDNA by using an iScript cDNA synthesis kit (Bio-Rad). cDNA was diluted with water at a ratio of 1:10, and 2.5 μl of the cDNA mixture was subject to qPCR with SYBR green I master mix and the LightCycler 480 system (Roche). Alternatively, cDNAs were generated by conventional techniques using Superscript II RT (Thermo-Fisher). Random hexamers were used for the majority of experiments to ensure detection of pre-mRNAs, nascent RNAs, and mature mRNAs. However, dT was used for cDNA synthesis in the experiments shown in Fig. 5B to E. Primers are listed in Table 1.
TABLE 1.
Primers used in this study
F, forward; R, reverse.
UV crosslinking and RNA-protein interaction.
RNAs associated with NS1 during infection or during transfection of NS1 were isolated as described previously (48). Briefly, A549 cells were infected with A/WSN/33 at an MOI of 5. At 6 hpi, cells were crosslinked with UV at 250 mJ/cm2 and lysed with SDS lysis buffer (0.5% SDS, 50 mM Tris-Cl [pH 6.8], 1 mM EDTA, 1 mM DTT, 2.5 mg/ml cRNA, 10 mM ribonucleoside-vanadyl complexes [VRC]). Immunoprecipitation was performed with anti-NS1 antibody or rabbit IgG. After a wash with RIPA buffer (1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-Cl [pH 8.0], 2 mM EDTA), associated RNA was treated with proteinase K at 37°C overnight and isolated with phenol-chloroform-isoamyl alcohol. RNA was transcribed into cDNA and analyzed by qPCR. For transfection experiments shown in Fig. 8, HEK293T cells were transfected with the indicated plasmids encoding the various NS1 proteins for 24 h. UV-RIP followed by RT-qPCR was performed as described above. The recovery efficiency (percentage of the input) of the RNA-IP was calculated as 2CT sample − CT input × 1%.
RNA-seq data processing and peak calling.
The RNA-seq data were tested for quality using FastQC (49). The sequence data were aligned to the human genome using Bowtie (50). PCR duplicates of the RNA-seq reads were removed. The differential peak calling of reads from experimental samples was compared to that of controls using modified PIPE-CLIP (23), using default cutoffs. Differential peaks were selected, and the corresponding genes are listed as hits.
RNA binding motifs were identified using the MEME-SUITE (51) motif discovery package with default search parameters. An enrichment cutoff P value of <0.01 was used to identify significant motifs. Identified motifs were then compared to previously documented RNA-binding protein (RBP) binding regions (52) and further annotated using the CISBP-RNA database (52).
Gene set enrichment analysis.
GSEA was performed by using the Canonical Pathways of MolecularSignatures Database (MSigDB) of the Broad Institute (53). Enriched gene sets (pathways) were sorted based on the false discovery rate (FDR) values.
Interferon-regulated genes and immune-related genes.
The databases for interferon-regulated genes and immune-related genes were obtained from the Interferome (http://www.interferome.org/) (54) and ImmPort (http://www.immport.org/) databases, respectively.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided by NIH grant R01 GM113874-01 to B.M.A.F. and N.K.C., NIH grant R01 AI125524-01 to B.M.A.F. and A.G.-S., Welch Foundation grant I-1915-20170325 (to N.K.C), NIH grant R01 AI123165 (to N.K.C.), grants from National Natural Science Foundation of China (81472725 to L.Z. and W.M. and 31872642 to L.Z.), and the Fundamental Research Funds for the Central Universities (20720160072) to L.Z. These studies were also partially supported by CRIP (Center for Research in Influenza Pathogenesis), an NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS) (contract number HHSN272201400008C) and by NIH grants U19AI106754 and U19AI117873 to A.G.-S.
L.Z., N.K.C., A.G.-S., and B.M.A.F. conceptualized the study; L.Z., N.K.C., M.K., A.G.-S., and B.M.A.F. performed methodology; L.Z., J.W., W.M., D.S., R.S., A.A., and R.M.-M. performed investigations; L.Z., N.K.C., and B.M.A.F. wrote the original draft; L.Z., N.K.C., A.G.-S., and B.M.A.F. wrote, reviewed, and edited the manuscript; B.M.A.F., N.K.C., and A.G.-S. acquired funding; A.G.-S., N.K.C., and B.M.A.F. provided resources; and L.Z., A.G.-S., N.K.C., and B.M.A.F. supervised the study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01634-18.
REFERENCES
- 1.Ayllon J, Garcia-Sastre A. 2015. The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- 2.Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci U S A 103:14194–14199. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuss-Duerkop SK, Wang J, Mena I, White K, Metreveli G, Sakthivel R, Mata MA, Muñoz-Moreno R, Chen X, Krammer F, Diamond MS, Chen ZJ, García-Sastre A, Fontoura BMA. 2017. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog 13:e1006635. doi: 10.1371/journal.ppat.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrhardt C, Wolff T, Pleschka S, Planz O, Beermann W, Bode JG, Schmolke M, Ludwig S. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J Virol 81:3058–3067. doi: 10.1128/JVI.02082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM. 2014. Viral subversion of nucleocytoplasmic trafficking. Traffic 15:127–140. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. EMBO J 18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3' end formation of cellular pre-mRNAs. Mol Cell 1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 8.Qiu Y, Krug RM. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J Virol 68:2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, Garcia-Sastre A, Roeder RG, Tarakhovsky A. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A 104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Krug RM. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4:55–64. [PMC free article] [PubMed] [Google Scholar]
- 12.Mor A, White A, Zhang K, Thompson M, Esparza M, Munoz-Moreno R, Koide K, Lynch KW, Garcia-Sastre A, Fontoura BM. 2016. Influenza virus mRNA trafficking through host nuclear speckles. Nat Microbiol 1:16069. doi: 10.1038/nmicrobiol.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 14.Bornholdt ZA, Prasad BV. 2008. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. 2011. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot P-E, Ciancia C, Perrin-Cocon L, Bartenschlager R, André P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog 9:e1003440. doi: 10.1371/journal.ppat.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo RL, Li ZH, Li LH, Lee KM, Tam EH, Liu HM, Liu HP, Shih SR, Wu CC. 2016. Interactome analysis of the NS1 protein encoded by influenza A H1N1 virus reveals a positive regulatory role of host protein PRP19 in viral replication. J Proteome Res 15:1639–1648. doi: 10.1021/acs.jproteome.6b00103. [DOI] [PubMed] [Google Scholar]
- 18.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Burckstummer T, Muller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G. 2012. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 19.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LCF, Yángüez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, König R, Stertz S, García-Sastre A, Chanda SK. 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Fu B, Li W, Patil G, Liu L, Dorf ME, Li S. 2017. Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat Commun 8:13876. doi: 10.1038/ncomms13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuss SK, Mata MA, Zhang L, Fontoura BM. 2013. Nuclear imprisonment: viral strategies to arrest host mRNA nuclear export. Viruses 5:1824–1849. doi: 10.3390/v5071824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, Yun J, Kim MS, Mendell JT, Xie Y. 2014. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol 15:R18. doi: 10.1186/gb-2014-15-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortes P, Beloso A, Ortin J. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J 13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu Y, Nemeroff M, Krug RM. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 26.Brugiolo M, Herzel L, Neugebauer KM. 2013. Counting on co-transcriptional splicing. F1000Prime Rep 5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastasina M, Le May N, Bugai A, Fu Y, Soderholm S, Gaelings L, Ohman T, Tynell J, Kyttanen S, Barboric M, Nyman TA, Matikainen S, Julkunen I, Butcher SJ, Egly JM, Kainov DE. 2016. Influenza virus NS1 protein binds cellular DNA to block transcription of antiviral genes. Biochim Biophys Acta 1859:1440–1448. doi: 10.1016/j.bbagrm.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol 77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrage F, Dutta K, Nistal-Villán E, Patel JR, Sánchez-Aparicio MT, De Ioannes P, Buku A, Aseguinolaza GG, García-Sastre A, Aggarwal AK. 2012. Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure 20:2048–2061. doi: 10.1016/j.str.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. 2013. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep 14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 34.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 37.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Mibayashi M, Martínez-Sobrido L, Loo Y-M, Cárdenas WB, Gale M Jr, García-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Aparicio MT, Ayllon J, Leo-Macias A, Wolff T, Garcia-Sastre A. 2017. Subcellular localizations of RIG-I, TRIM25, and MAVS complexes. J Virol 91:e01155-16. doi: 10.1128/JVI.01155-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res 31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 41.Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- 42.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD, Wherry EJ. 2011. A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog 7:e1002098. doi: 10.1371/journal.ppat.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. 2006. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 107:2613–2618. doi: 10.1182/blood-2005-07-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Das P, Schmolke M, Manicassamy B, Wang Y, Deng X, Cai L, Tu BP, Forst CV, Roth MG, Levy DE, Garcia-Sastre A, de Brabander J, Phillips MA, Fontoura BM. 2012. Inhibition of pyrimidine synthesis reverses viral virulence factor-mediated block of mRNA nuclear export. J Cell Biol 196:315–326. doi: 10.1083/jcb.201107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai PL, Chiou NT, Kuss S, Garcia-Sastre A, Lynch KW, Fontoura BM. 2013. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog 9:e1003460. doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conrad NK. 2008. Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Methods Enzymol 449:317–342. doi: 10.1016/S0076-6879(08)02415-4. [DOI] [PubMed] [Google Scholar]
- 47.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 48.Sei E, Conrad NK. 2014. UV cross-linking of interacting RNA and protein in cultured cells. Methods Enzymol 539:53–66. doi: 10.1016/B978-0-12-420120-0.00004-9. [DOI] [PubMed] [Google Scholar]
- 49.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Institute, Cambridge, United Kingdom: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- 50.Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11:Unit 11.17. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.