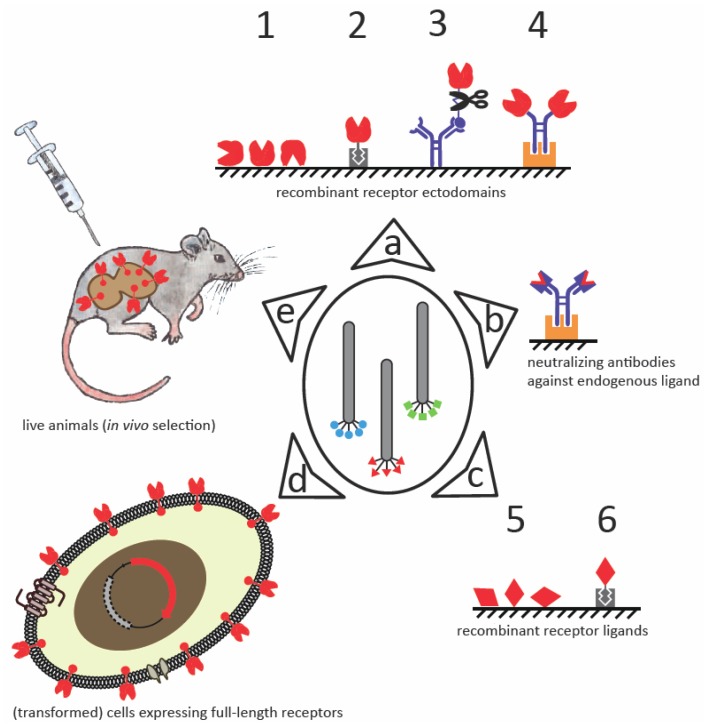
Figure 1.
Schematic representation of some well-established biopanning approaches to identify membrane receptor-binding peptides from phage-displayed libraries. (a) Due to the difficulties accompanying isolation and recombinant expression of whole membrane receptors, only the extracellular regions (shown in red) or their individual domains can be used as target molecules. These can be immobilized to an appropriate matrix (oblique striped grounding) by direct adsorption (1) or by a suitable affinity tag that is recognized by its respective binding partner (e.g., biotin-avidin interaction, 2) or a specific antibody (3). A linker connecting the affinity tag to the receptor fragment that contains a specific protease cleavage site (3) can be introduced to facilitate the specific elution of target-bound phages by protease treatment. Alternatively, receptors may be imitated by recombinant antibody chimeras (receptor ectodomains fused to the Fc-region of an antibody; 4), in which case the target molecule is specifically captured to protein A- or G-coated matrix (shown in orange). (b) Targeting neutralizing antibodies against the endogenous ligand can result in selection of peptides binding to cognate receptor. (c) The ligand-receptor interaction can be inhibited by targeting the endogenous ligand instead of the receptor. Again, direct (5) or indirect immobilization (6) of the ligand is possible. (d) A more advanced strategy is to screen against whole cells over-expressing the full-length membrane-embedded receptor; this is usually achieved by transforming cells with a gene encoding the receptor. (e) In the in vivo approach, library phages are injected intravenously into an animal, and specific binders are recovered from tissue biopsies. The identities of the targeted receptors are determined afterwards.