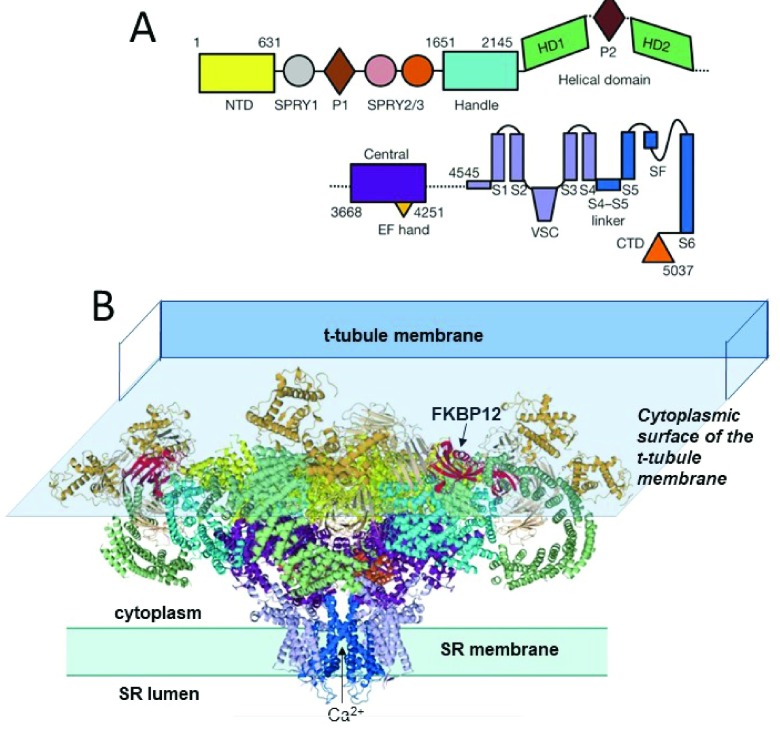
Figure 1. The near-atomic resolution structure of RyR1 10.
( A) The linear sequence of RyR1 domains, starting at the N-terminal domain (NTD). ( B) “Side” view of RyR1 highlighting two of four protomers. Domains in ( B) are colour-coded in the same way as those in ( A). Modified from Figure 1 of Yan et al. 10. The 10 major domains in each subunit include the N-terminal domain, which harbours many RyR1 and RyR2 disease-causing mutations. They also include the SPRY1 domain that forms part of the binding site for FKPB12, the helical and central domains, which contain protein kinase A and Ca 2+/calmodulin protein kinase II (CaMKII) phosphorylation sites, the transmembrane domain containing the ion pore, and the C-terminal domain that forms part of the Ca 2+, ATP, and caffeine activation sites 17 ( Figure 2). FKBP12 binds in a cleft formed by the Handle, NTD, and SPRY1/3 domains. Suggested FKBP12 binding residues are located in the Handle domain (P1780, C1781, and S1687) 10 and in a hydrophobic cluster around D720 in the SPRY1 domain 26. The cytoplasmic surface of the transverse tubule (T-tubule) membrane is shown overlying the RyR to illustrate the way that the bulk of the RyR is sandwiched between the membranes on either side of the T-tubule/sarcoplasmic reticulum (SR) junction. RyR, ryanodine receptor.