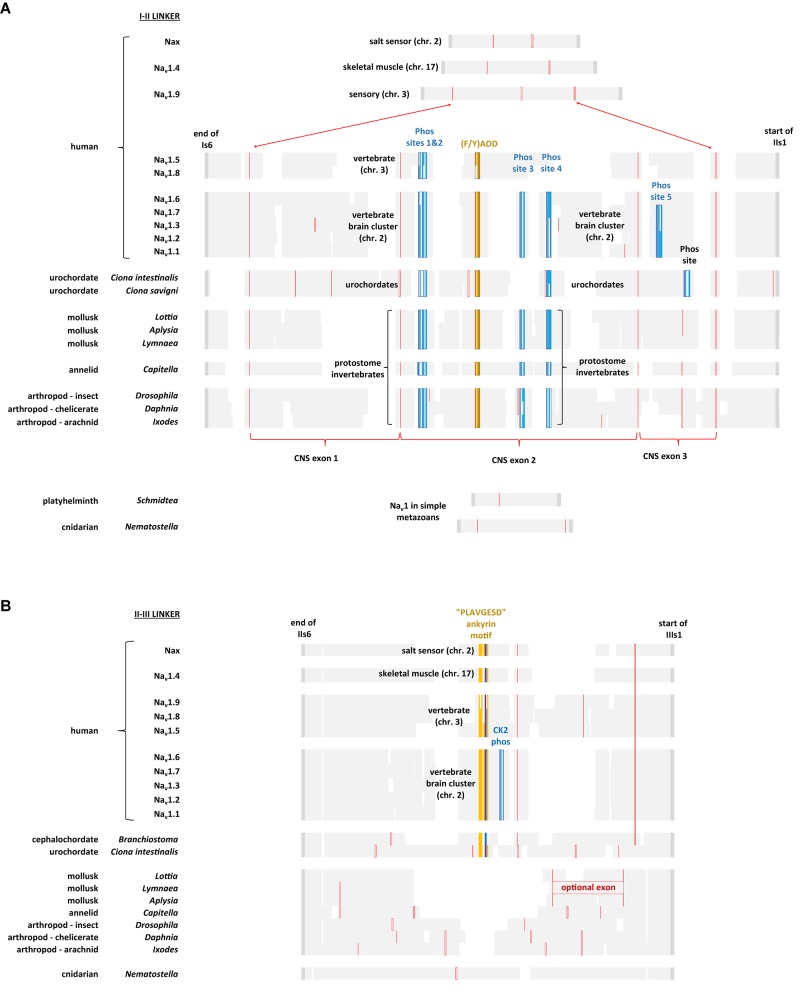
FIGURE 11.
Alignment of amino acid sequences coding for cytoplasmic I-II linker (A) and II-III linker (B) of invertebrate and vertebrate Nav1 channels. Note the expanded size of the cytoplasmic I-II linker in protostome invertebrate Nav1 and vertebrate brain cluster channels (Nav1.x, where x = 1,2,3,6, and 7) containing four conserved phosphorylation sites at serine residues. Two of these four phosphorylation sites are subject to phosphorylation in mammalian Nav1.2 channels by cAMP dependent protein kinase A (serine: 573 and 623) (Smith and Goldin, 1996) and one of the four is a protein kinase C regulatory site (serine 576) (Cantrell et al., 2002). Other vertebrate sodium channels, such as Nav1.5 and Nav1.8 retain two out of four of these phosphorylation sites (serine: 573 and 576) while Nav1.4, Nav1.9 and Nax lack any of these phosphorylation sites. While all chordate Nav1 channels possess the ankyrin G binding motif, only the nervous system specific cluster of vertebrate sodium channels (Nav1.1, Nav1.2, Nav1.3, Nav1.6, and Nav1.7) possess a CK2 serine/threonine kinase phosphorylation site downstream of the ankyrin G binding site in the II-III linker, that greatly increases affinity and required for the formation of stable ankyrin G – sodium channel complexes upon phosphorylation (Xu and Cooper, 2015). Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).