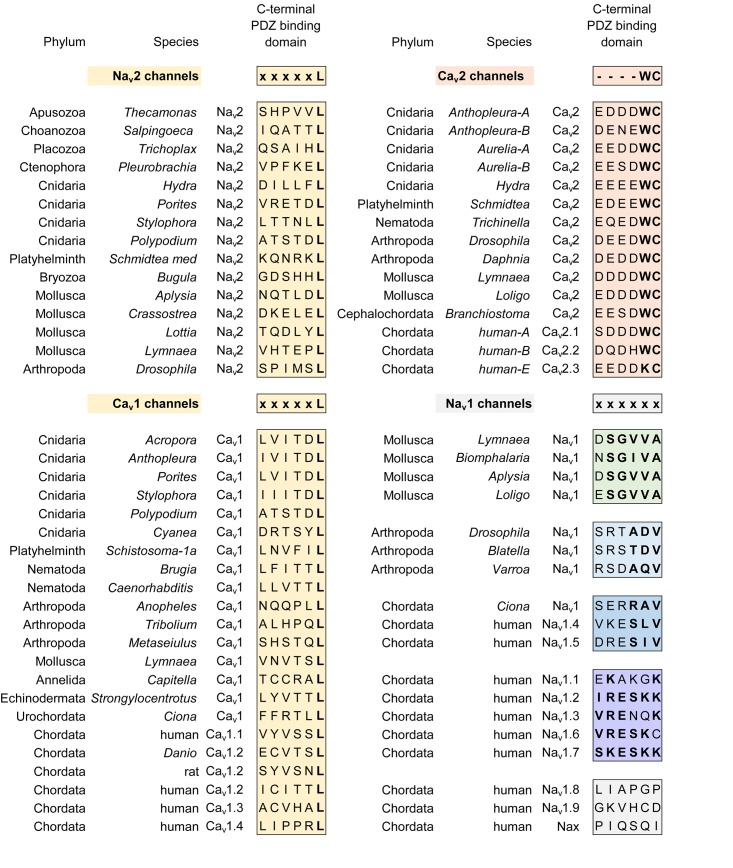
FIGURE 12.
Alignment of amino acid sequences coding for the C-terminal ends of calcium channels (Cav1 and Cav2) and sodium channels (Nav2 and Nav1) where there are conserved sequence motifs associated with binding to different classes of PDZ domain containing proteins. There is a surprising consistency in a xxxxL motif conserved between Nav2 and Cav1 channels (yellow color), both of which constitute the basal complement of high voltage-activated calcium channels and sodium channels present in genomes of single cell choanoflagellates such as S. rosetta. The Nav1 and Cav2 channels, which are likely derived from Nav2 and Cav1 channels, respectively, possess different sets of PDZ binding domain motifs (denoted by orange, green, blue, purple colors). PDZ binding motifs uniquely diverge within the Nav1 channel class where molluscan, arthropod, and different human sodium channels associate with separate classes of PDZ domain containing proteins, contributing to their specific localization within their unique cellular environments. The “SLV/SIV” C-terminal PDZ binding motif in mammalian Nav1.4 and Nav1.5, for example, aids in the complexing of these Nav1 channels to syntrophins and SAP97, for the cellular targeting within skeletal and cardiac muscle, respectively. Cav3 channels lack conservation in a C-terminal PDZ domain. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).