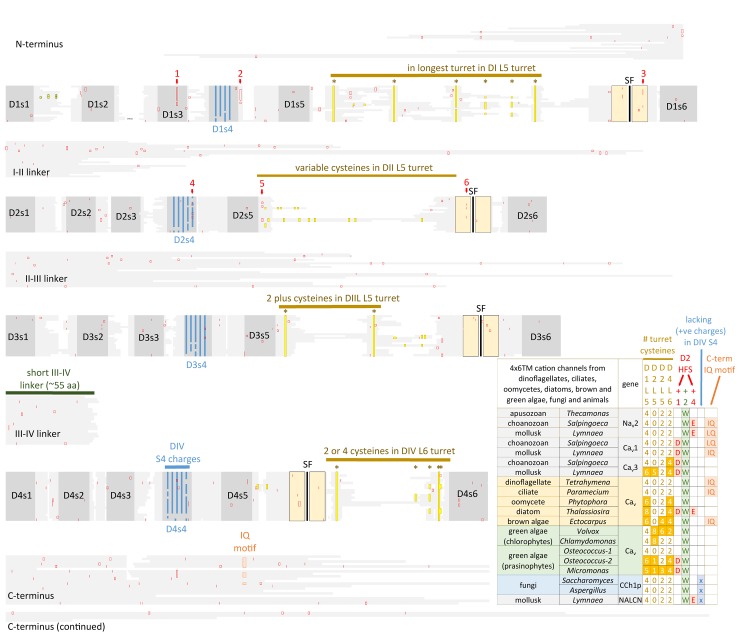
FIGURE 7.
Alignment of 4 × 6 TM cation channel representatives illustrating a shared fundamental architecture of asymmetrically arranged extracellular and intracellular regions from single cell protists (choanoflagellates, dinoflagellates, oomycetes, and ciliates), green algae (chlorophytes and prasinophytes), brown algae, yeast, and Nav2, Nav1, Cav1, Cav2, and Cav3 channels from protostome invertebrate (pond snail, L. stagnalis). A concerted evolutionary change to a common asymmetrical architecture of 4 × 6 TM cation channels contrasts with the pseudo-symmetry of cyclic nucleotide-gated potassium channel (CNGK) channels which contain four nearly identical domains chained together. Long extensive extracellular turrets prior to the pore selectivity filter (SF) include L51, L5III and more rarely L5II and a longer turret after the SF in L6IV, as well as a 4-0-2-2 framework of core cysteines populating L5I-L5II-L5III-L6IV extracellular turrets in 4 × 6 TM channels. L5IV extracellular turret is always short in 4 × 6 TM channels. The cytoplasmic III-IV linkers are always short (∼53–57 amino acids) compared in contrast to more variable I-II linker, II-III linkers, and N- and C-termini in 4 × 6 TM cation channels. Intron splice sites are indicated by red vertical lines. Dinoflagellate, ciliate, oomycete, diatom, green and brown algae channels illustrated in the figure all possess a conserved HFS site resembling an animal calcium channel (EEEE). Notably lacking in most of these protist channels is the conservation in the pore selectivity filter of Domain II of a “calcium beacon” aspartate (D) (HFS+1) omni-present in animal calcium channels, or a “sodium beacon” glutamate (HFS+4) omni-present in animal sodium channels, and a C-terminal IQ motif. A tryptophan ring at HFS+2 is present in all four domains of all channels except Domain I of Cch1p. A notable absence of S4 charges is found for yeast Cch1p and NALCN. Amino acid sequences were aligned using MUSCLE 3.7 (Edgar, 2004) within EMBL-EBI web interface (Chojnacki et al., 2017).