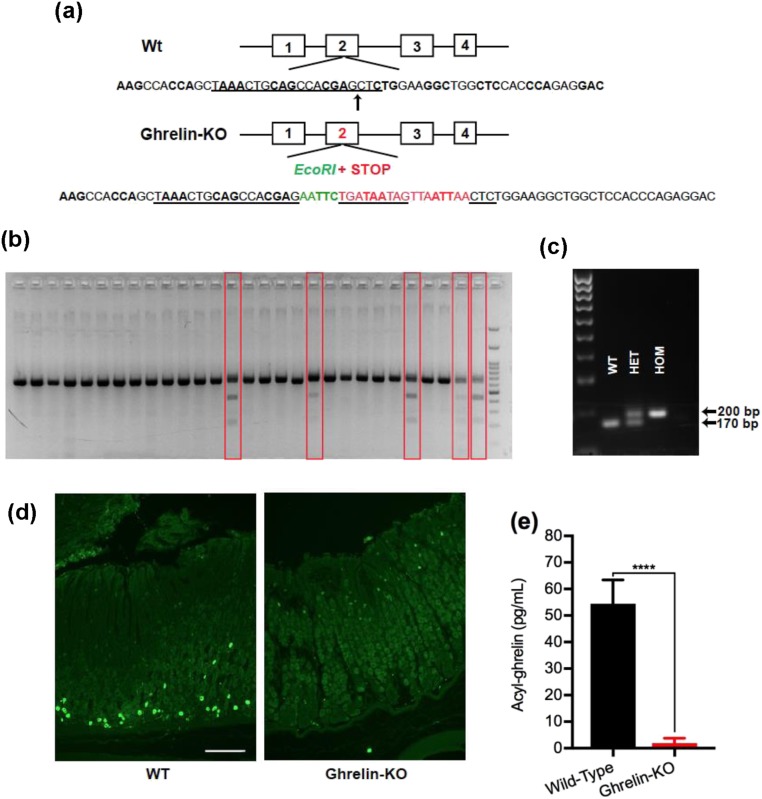
Figure 1.
Generation and validation of the novel ghrelin-KO mouse lines GKO1 and GKO3. (a) Schematic of the derivation of ghrelin-KO mice using CRISPR-Cas9 genome editing to insert an EcoR1 site (to facilitate screening for the edited gene) and multiple STOP codons into exon 2 of the Ghrl gene (at the insertion site marked by an arrow). The sequence targeted by the short guide RNA appears in black with underlining. The green-colored nucleotides represent the inserted EcoR1 sequence. The red-colored nucleotides with underlining represent three sequential in-frame STOP codons. The red-colored nucleotides without underlining indicate additional out-of-frame STOP codons. (b) Gel image showing separation of genotyping PCR products from potential ghrelin-KO founders (red boxes) and nontargeted mice, after EcoR1 digestion. (c) PCR analysis of genomic DNA obtained by tail biopsies of representative mice derived from crosses of mice heterozygous for the ghrelin-KO allele, demonstrating identification of mice with two copies of the wild-type Ghrl allele (WT), mice homozygous for the knockout Ghrl allele (HOM), and heterozygotes (HET). (d) Immunohistochemistry of representative stomach sections from 3- to 4-mo-old WT and ghrelin-KO littermates of the GKO3 line. The green color represents ghrelin immunoreactivity. Scale bar, 100 μm. (e) Plasma acyl-ghrelin levels from 28-d-old nonfasted WT and GKO3 littermates. n = 9 to 10 (combination of males and females). Data are expressed as mean ± SEM. ****P < 0.0001, by a Student unpaired t test.