Fig 2.
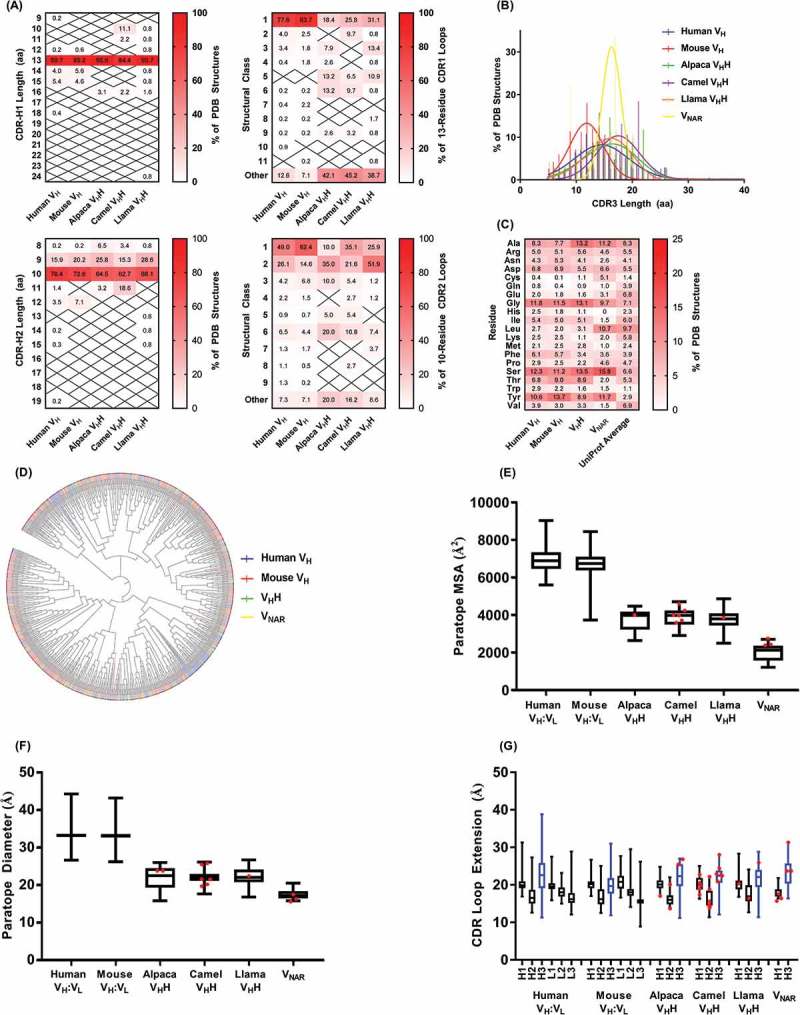
Properties of sdAb vs. conventional antibody paratopes. (A) Structural classification of CDR1 and CDR2 according to PyIgClassify.108 (B) CDR3 length distributions. (C) Amino acid compositions of conventional antibody (VH domain) and sdAb paratopes. For VHHs, sequences of CDR1, CDR2 and CDR3 (Honegger-Plückthun numbering) were used and for VNARs, sequences of CDR1 and CDR3 only were used. (D) Relatedness of conventional antibody (VH domain) and sdAb CDR3 sequences. The phylogenetic tree was produced using neighbor-joining methods in ClustalW2 and the cladogram was visualized using iTOL109 with CDR3s colored according to species origin as in part B. (E) Molecular surface areas of conventional antibody (VH:VL) and sdAb paratopes. Areas were calculated for merged CDR sequences (Honegger-Plückthun numbering) using PyMol. (F) Diameters of conventional antibody (VH:VL) and sdAb paratopes. Diameters were calculated as the maximum interatomic distance between any two FR-CDR boundary residues (Honegger-Plückthun numbering). (G) Extension of conventional antibody (VH:VL) and sdAb paratopes. Extension was calculated as the maximum interatomic distance between the CDR base (first or last residue according to Honegger-Plückthun numbering) and the CDR tip. The CDR(H)3 loop is shown in blue. In parts (E) – (G), boxplot lines represent medians, the box boundaries represent quartiles and the box whiskers represent ranges. Red dots indicate sdAbs targeting cryptic epitopes discussed in the main text. Data are representative of all complete antibody structures available in the Protein Data Bank and indexed in PyIgClassify as of January 2018.
