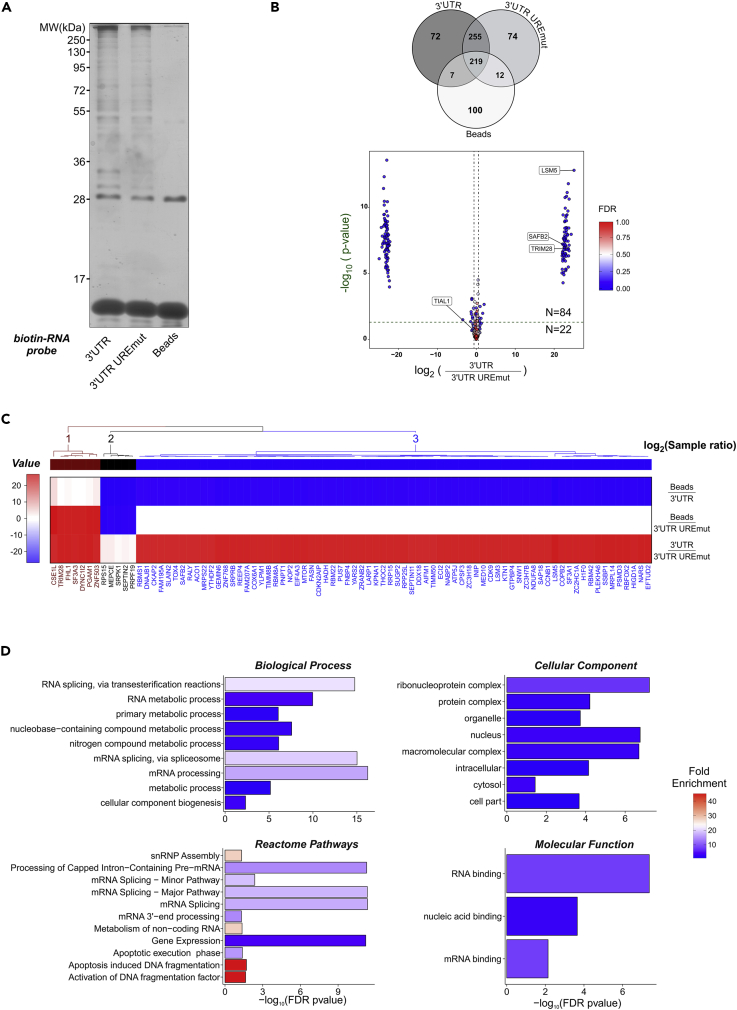
Figure 4.
Identification of Proteins Binding to the UREs within the Human BOK 3’ UTR
(A and B) HEK293T lysates were incubated with biotinylated RNA probes encoding human BOK CDS-3’UTR or CDS-3’UTR(UREmut1-3), a version with the three predicted URE sites mutated. Pull-down fractions were analyzed for the presence of RNA-binding proteins by SDS-PAGE and silver staining (A) and shotgun tandem mass spectrometry (MS/MS). The proteins detected were represented with a Venn diagram reflecting the count of detected proteins per sample type (B upper panel) and a volcano plot with the ratio and statistical significances shown between the indicated probes (B lower panel). Vertical (magenta) and horizontal (green) dashed lines mark the thresholds for absolute fold change (≥±1.5) and p values (p ≤ 0.05). A continuous color scale is applied to each point in accordance to the estimated false discovery rate (FDR <1%). Values corresponded to averages from three independent experiments.
(C) Clustering of the paired sample ratio abundances for the identified hits.
(D) A total of 106 candidates with a positive fold change (≥1.5) were analyzed in the database Panther (http://pantherdb.org/) for overrepresentation within four gene ontology categories performing a Fischer test with FDR correction. Bars represent the -log10-transformed corrected p value and are filled according to the calculated fold enrichment against the corresponding human genome background.