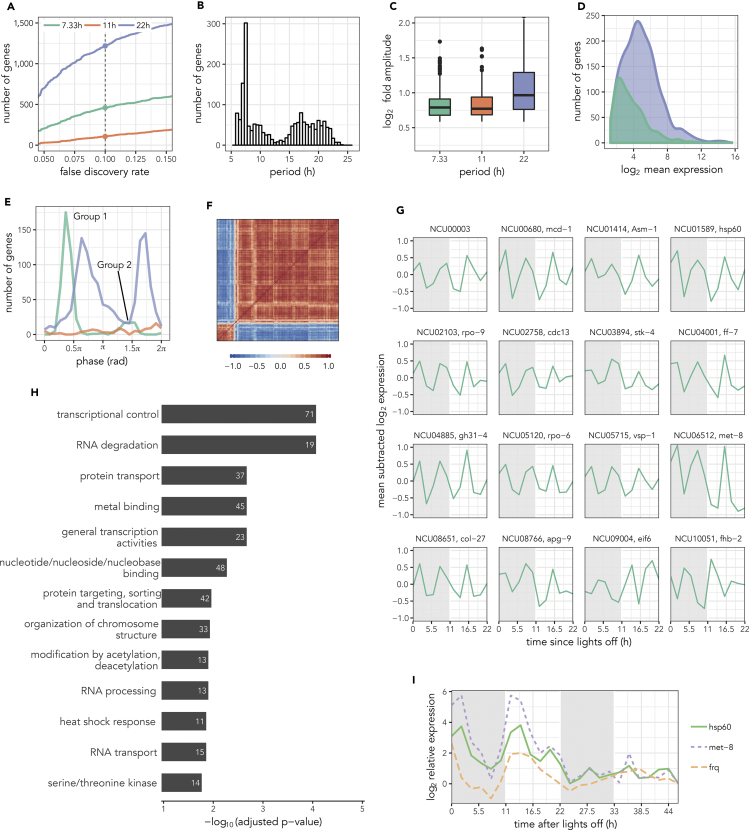
Figure 1.
Quantification of 7-hr Transcripts in N. crassa
(A–I) (A) The number of rhythmic genes at each period identified by the model-selection-based procedure at different false discovery rate thresholds. The color coding for the different periods is maintained throughout the figure. (B) The distribution of periods of the rhythmic genes as estimated by the ARSER (Wu et al., 2016). The distribution of peak-to-peak fold amplitudes (C), mean expression level (D), and phases (E) of the different rhythmic genes quantified by the model-selection-based procedure. The second harmonic genes have been omitted in (D) for clarity. (F) Correlation-metric-based hierarchical clustering of the mean-subtracted expression profiles of the third harmonic genes. (G) Transcript profiles of selected third harmonic genes, such as transcription factors, kinases, and chromatin remodelers. (H) Top functional categories (FDR<0.05) enriched in the third harmonic genes using FungiFun2 (Priebe et al., 2015). (I) Verification of 7-hr rhythms in two candidate genes using quantitative real-time PCR over a 2-day time course with 2-hr time resolution. The results for met-8 and hsp60 in a WT strain are shown. The circadian clock gene frq is included as a positive control of the quantitative real-time PCR analysis. Expression levels relative to the last time point are plotted for each gene. See also Figures S1 and S2 and Transparent Methods.