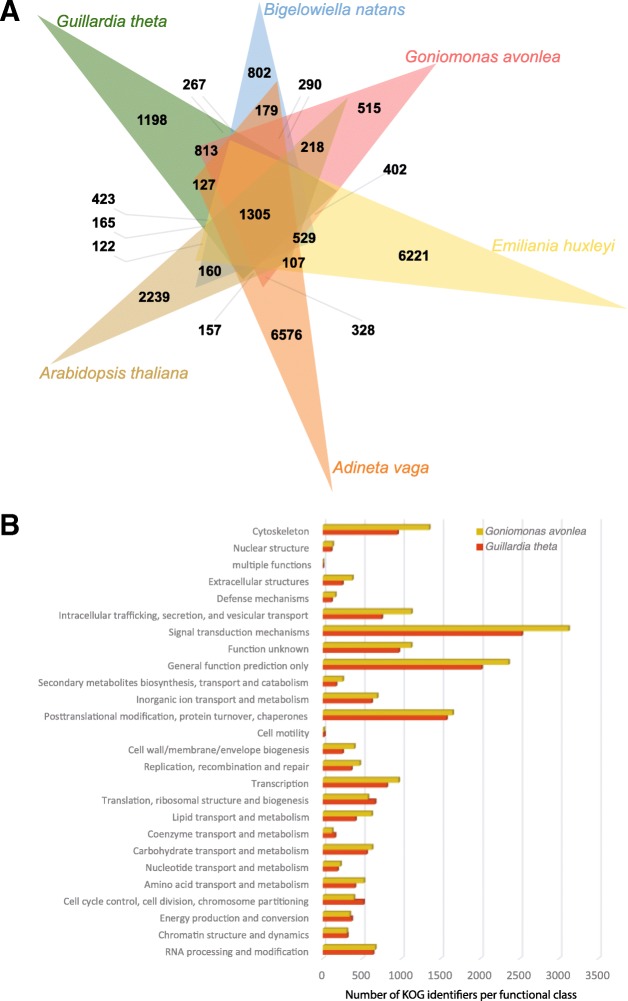
Fig. 1.
Comparative genomics of Goniomonadea, Cryptophyceae, and other eukaryotes. a Venn diagram showing orthologous clusters shared between the goniomonad Goniomonas avonlea (red), the cryptophyte Guillardia theta (green), the rhizarian Bigelowiella natans (blue), the haptophyte Emiliania huxleyi (yellow), the opisthokont Adineta vaga (orange), and the land plant Arabidopsis thaliana (brown). Go. avonlea shares 4321 families with Gu. theta, higher than is shared with other eukaryotes (B. natans (3647), E. huxleyi (3441), A. vaga (3173), A. thaliana (2955)). b KOG classification of proteins in Go. avonlea (brown) and Gu. theta (red). Within most functional categories, the number of proteins in the two organisms is similar. However, Go. avonlea possesses more proteins in some categories, in particular the cytoskeleton and the intracellular trafficking, secretion, and vesicular transport families