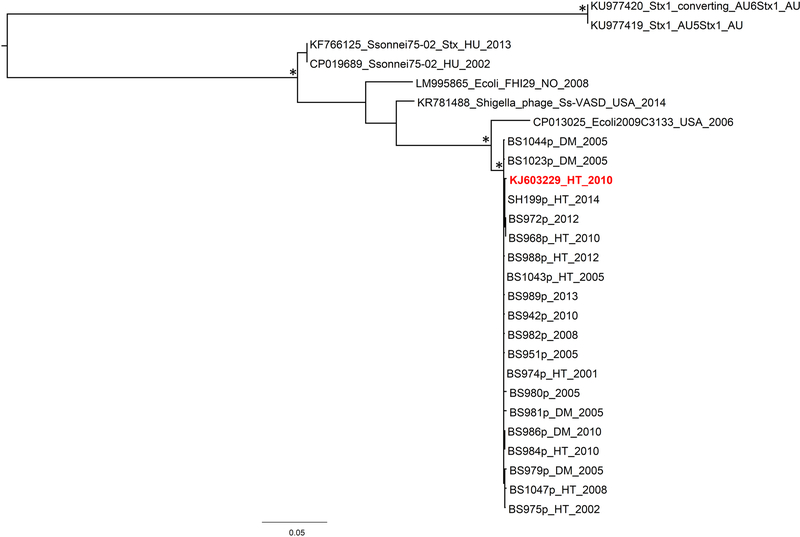
Figure 2. Maximum Likelihood tree inferred on the alignment of unique phage sequences from Hispaniola plus reference sequences from GenBank.
The reference sequence of Shigella phage φPOCJ13 (GenBank accession no. KJ603229) is reported in red. BS984 and BS986 are phages extracted from S. boydii 19 strains, while strains BS975, BS979 and BS1047 are extracted from S. dysenteriae 4 strains. All the other phages are encoded in S. flexneri strains. The country of origin and time of isolation (when known) are indicated in the label of each strain by a two/three letter code (AU = Australia, DM = Dominican Republic, HT= Haiti, HU = Hungary, NO = Norway, USA = United States) followed by the year. The tree was rooted using the divergent Australian phage sequences from E. coli as an outgroup. Branch lengths are drawn to scale proportional to SNPs distance per SNP site according to the scale bar at the bottom of the tree. Ultrafast bootstrap support >90% for internal branches of the tree is indicated by an asterisk.