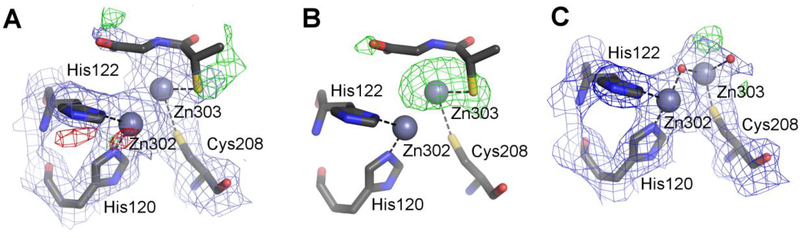
Figure 2.
Various stages of modeling chain A of the structure of NDM-1 5a5z (PDB ID of the re-refined model: 5nbk). All maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). (A) The original coordinates with the electron density maps produced by REFMAC5 without prior refinement, with the putative tiopronin ligand visible at the top. (B) The original coordinates but with the omit map calculated from the final refinement phases by omitting only the water molecules modeled in the active site. (C) The re-refined entry 5nbk, with the final electron density maps. The ligands and the electron density maps, including omit maps, can be inspected using an interactive figure created with Molstack (http://molstack.bioreproducibility.org/project/view/2gniVO4HvvzPJnvb0Sd7/).