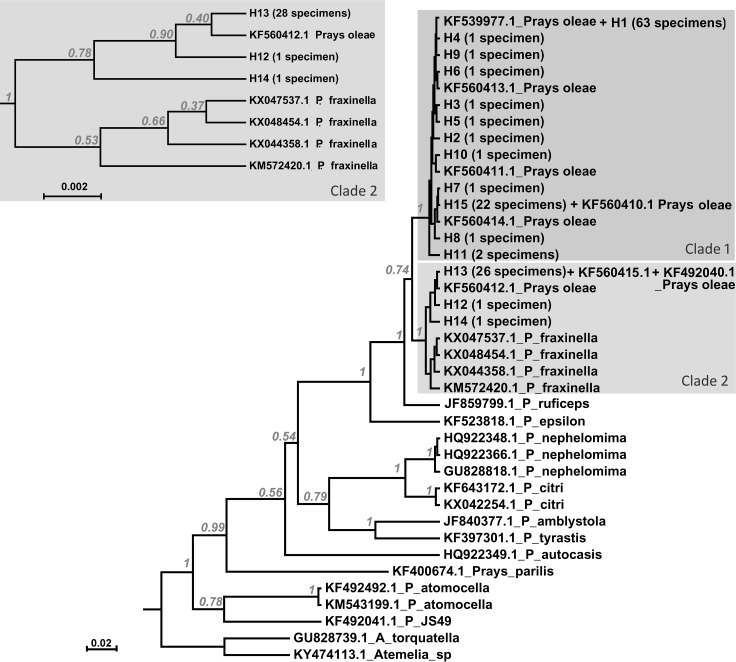
Fig 2. Phylogenetic relationship between Prays oleae and other Prays species with data available on GenBank (accession code given on tree), based on the COI amplicon.
The phylogeny corresponds to the majority rule consensus tree of trees sampled in a Bayesian analysis, and the posterior probability values are shown for main nodes (xml input files available in S1 File). Two Atemelia species were used as outgroups. Similar topologies were obtained using Maximum Likelihood and Neighbor-Joining methods as implemented in Mega 7.0 [19] (S1 and S2 Figs) and will further not be discussed. The top left grey window highlights a clade showing Prays oleae specimens nesting in the Prays fraxinella clade, an unexpected result. Prays oleae samples were collected from Portugal with exception of KF560413.1, KF560414.1, and KF560415.1 which were collected in Tunisia and KF492040.1 from Spain. The tip labels represent either the haplotypes and number of specimens with that same haplotype (details in S2 Table) or the GenBank accession number.