Abstract
Ribosomes perform protein synthesis but are also involved in signaling processes, the full extent of which are still being uncovered. We report that phenotypes of mutating ribosomal proteins (Rp’s) are largely due to signaling. Using Drosophila, we discovered that a bZip-domain protein Xrp1 becomes elevated in Rp mutant cells. Xrp1 reduces translation and growth, delays development, is responsible for gene expression changes, and causes the cell competition of Rp heterozygous cells from genetic mosaics. Without Xrp1, even cells homozygously-deleted for Rp genes persist and grow. Xrp1 induction in Rp mutant cells depends on a particular Rp with regulatory effects, RpS12, and precedes overall changes in translation. Thus effects of Rp mutations, even the reduction in translation and growth, depend on signaling through the Xrp1 pathway and are not simply consequences of reduced ribosome production limiting protein synthesis. One benefit of this system may be to eliminate Rp-mutant cells by cell competition.
Keywords: ribosomal protein, Drosophila development, cell competition, Xrp1, regulation of translation, growth regulation, Minute mutation, ribosomopathy
eTOC
Certain mutant cells are eliminated only in mosaic tissues. Lee et al. isolate mutations in a bZip domain protein, Xrp1, that rescue ribosomal protein (Rp) heterozygous mutant cells from such competition. Xrp1 accounts for many aspects of the Rp mutant phenotype, indicating they are not direct consequences of ribosome depletion.
INTRODUCTION
Ribosomes are the essential protein synthetic machines of the cell. Large and small subunits (LSU and SSU), 40S and 60S in eukaryotic cells, form an 80S complex together with mRNA and perform translation in the cytoplasm. Each ribosome subunit is a ribonucleoprotein complex containing one (SSU) or three (LSU) non-coding rRNA molecules and a battery of ribosomal proteins (Rp’s) and is assembled in the nucleolus for export to the cytoplasm. Ribosomal proteins can contribute to folding and assembly of the ribosomal subunits as well as their function in translation(de la Cruz et al., 2015). Most ribosomal proteins are essential and cells homozygous for their mutations die, while heterozygous Rp mutants that lack one copy of the gene are abnormal in both humans and in Drosophila.
To what extent do the defects in ribosomal protein mutants reflect deficient translation, and to what extent do they reflect signaling pathways that monitor ribosome status? Aspects of Diamond Blackfan Anemia, the ribosomopathy that occurs in humans heterozygous for mutations in a number of Rp genes, are thought to reflect chronic p53 signaling, activated by accumulation of a ribosome assembly intermediate and nucleolar stress(Ellis, 2014; Raiser et al., 2014). On the other hand, Diamond Blackfan Anemia is also characterized by short stature and delayed maturation as well as skeletal defects (Vlachos et al., 2014; Mirabello et al., 2017), and Diamond Blackfan Anemia has sometimes been treated with L-leucine to stimulate protein synthesis (Pospisilova et al., 2007; Payne et al., 2012). Reduced protein synthesis has been measured in both Drosophila embryos and in mouse fibroblasts and hematopoietic cells from heterozygous, Rp+/− genotypes (Boring et al., 1989; Oliver et al., 2004; Cmejlova et al., 2006; Signer et al., 2014).
We made use of Drosophila to investigate the effects of Rp mutations further. Drosophila that are haploinsufficient for any of 66 of the 79 Rp genes exhibit a common phenotype first recognized a century ago (the ‘Minute’ phenotype) which includes a reduction in the size and thickness of bristles on the adult body (“Minute” bristles) and a developmental delay associated with reduced translation and growth rate (Bridges and Morgan, 1923; Boring et al., 1989; Marygold et al., 2007). Unlike the bristle structures, most mutant cells are of normal size, as are mutant flies themselves, suggesting that the extended growth period is sufficient to compensate for reduced cellular growth(Neufeld et al., 1998; Montagne et al., 1999). In fact mutant organs can be larger than normal, depending on the particular balance of growth between organs(Lin et al., 2011).
In Drosophila, and possibly in mammals, Rp+/− genotypes are subject to ‘cell competition’ in genetic mosaics(Morata and Ripoll, 1975; Oliver et al., 2004). If growing imaginal discs (progenitor cells that grow in an undifferentiated state in the larva to give rise to the adult tissues) contain both wild type and Rp+/− cells, the latter are progressively lost during growth. Conversely, wild type cells growing in Rp+/− backgrounds come to dominate developmental compartments at the expense of the Rp+/− cells (Morata and Ripoll, 1975; Simpson, 1979). Both competitive situations are associated with selective apoptosis of Rp+/− cells in proximity to wildtype, which is responsible for the loss of Rp+/− clones (Claveria and Torres, 2016; Baker, 2017). There are other genotypes that can be competed from genetic mosaics but neither is it clear that the mechanisms are the same nor whether deficits in translation or growth are required (Claveria and Torres, 2016; Baker, 2017). There are also examples of ‘super-competitor’ genotypes that can eliminate nearby wild type cells, even though wild type cells should have normal ribosomes(de la Cova et al., 2004; Moreno and Basler, 2004; Tyler et al., 2007; Neto-Silva et al., 2010). In the mouse embryo, cells expressing more Myc or less p53 are super-competitors (Claveria et al., 2013; Dejosez et al., 2013; Zhang et al., 2017).
Cell competition is also seen in mammalian cell co-cultures, in many cases eliminating hyperplastic or preneoplastic cells(Hogan et al., 2009; Kajita et al., 2010; Chiba et al., 2016; Wagstaff et al., 2016). Such cells can also be eliminated from mosaics with otherwise normal tissues in vivo(Leung and Brugge, 2012; Brown et al., 2017).
The studies reported here originated in a genetic screen designed to identify new components of cell competition. This led to isolation of a mutation affecting a bZip-domain protein gene, Xrp1 (Lee et al., 2016). Xrp1 was previously known as a putative transcription factor induced by p53 following X-irradiation of Drosophila, and implicated in genome maintenance, although no point mutant alleles had been studied previously(Brodsky et al., 2004; Akdemir et al., 2007). Xrp1 was also characterized as a component of the protein complex that binds to the P element transposon in Drosophila, and found to contribute to P element transposition(Francis et al., 2016). We report a major role for Xrp1 in multiple features of Rp mutants. Xrp1 expression is induced in mutant cells by a signal from the ribosome and controls cellular translation rate and growth in addition to cellular competitiveness and almost the entire gene expression signature of Rp+/− cells. Xrp1 is even responsible for eliminating cells homozygously mutant for essential Rp genes that are deficient for new ribosome biogenesis. We conclude that Xrp1 controls a cellular stress pathway that monitors ribosomal proteins, regulates multiple cellular properties and acts upstream of the major defects in global translation, which are in fact only indirectly related to the initial mutation of a ribosomal protein gene.
RESULTS
Xrp1 is required for cell competition
A point mutation in gene encoding the putative transcription factor Xrp1 was isolated in a genetic screen designed to reveal defects in cell competition (Lee et al., 2016). The Xrp1 transcription unit on the right arm of the third chromosome encodes two protein isoforms (Figure 1A). The Xrp1m2-73 allele we isolated corresponds to a G-to-T transition replacing a Glu codon with a premature stop codon(Lee et al., 2016). The mutation affects an exon common to both Xrp1 isoforms of the protein and is predicted to truncate the long form after Thr367 and the short form after Thr105, before an AT-hook motif and basic leucine zipper (bZip) domain that occur near the C-termini of both isoforms (Figure 1A).
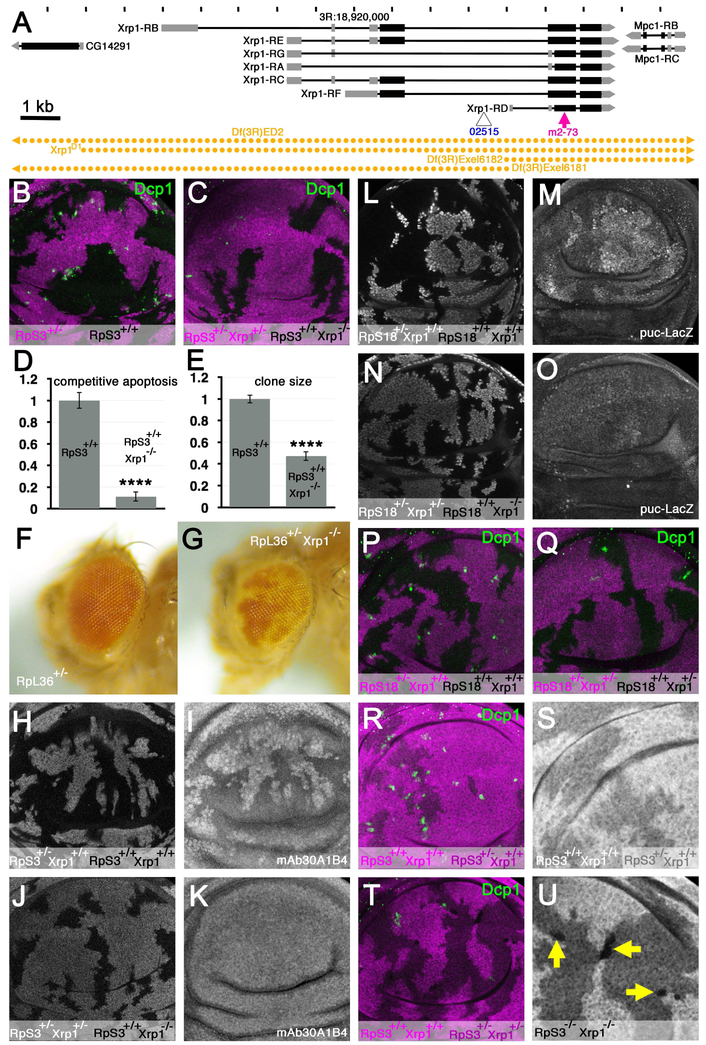
Figure 1. Xrp1 is required for cell competition.
A) Features of chromosome 3R around position 18.92 Mb. Seven Xrp1 transcripts encoding two proteins are encoded in a 14 kb region between the CG14291 and Mpc1 genes. Locations of the LacZ enhancer trap insertion Xrp102515 and truncation allele Xrp1m2-73 are shown. DNA segments absent from deletion strains used in this paper are indicated in gold. B) Cell competition affects RpS3+/− cells in wing discs containing RpS3+/+ clones. Dying cells labeled for active Dcp1 (green) are predominantly at the interfaces between RpS3+/− cells (magenta) and wild type cells (nor magenta). C) In the presence of an Xrp1 mutation, competitive cell death was reduced and clones of RpS3+/+ cells (unlabelled) colonized less of the tissue. See panels D-E for quantification. D) Competitive apoptosis of RpS3+/− cells next to RpS3+/+ cells was reduced ~10× in the presence of an Xrp1 mutation (p<0.0001, t-test)(in this case the RpS3+/− wing was also Xrp1+/−). Error bars are ± 1 S.E.M. E) Colonization of the RpS3+/− wing disc by wild type clones was reduced >2× by an Xrp1 mutation (p<0.0001, t-test)(the RpS3+/− wing was also Xrp1+/−). Error bars are ± 1 S.E.M. F) RpL36+/− clones (white) are rarely recovered in RpL36+/+ eyes(Tyler et al., 2007). G) RpL36+/− Xrp1−/− clones (white) were readily recovered in RpL36+/+ Xrp1+/− eyes. H, I) mAb30A1B4 detected an antigen that is expressed at higher levels in RpS3+/− cells in mosaic wing imaginal discs. J,K) mAb30A1B4 labeling of RpS3+/− Xrp1+/− cells dropped to the same level as RpS3+/+ Xrp1−/− cells in mosaic wing imaginal discs. L,M) Puc-LacZ was elevated in RpS18+/− cells compared to RpS18+/+ cells in mosaic wing discs. N,O) Puc-LacZ levels were reduced in RpS18+/− Xrp1+/− cells to levels similar to RpS18+/+ Xrp1−/− cells. P) Cell competition affected RpS18+/− cells in wing discs containing RpS18+/+ clones. Note that the reciprocal clone genotype RpS18−/− was not observed. Q) Competitive apoptosis of RpS18+/− cells was absent from mosaic wing discs containing RpS18+/+ clones when all cells were heterozygous for Xrp1. Note that the reciprocal clone genotype RpS18−/− was not observed. R) Cell competition affected RpS3+/− cells in wing discs containing RpS3+/+ clones (in this experiment, RpS3+/+ clones were positively marked with an extra dose of beta-galactosidase transgene (magenta). S) Enlargement of the beta-galactosidase channel from panel R. Note absence of any RpS3−/− cells lacking beta-galactosidase. T) Cell competition did not affect RpS3+/− Xrp1+/− cells in wing discs containing RpS3+/+ Xrp1+/+ clones. Competitive apoptosis was reduced and colonization by RpS3+/+ Xrp1+/+ cells was reduced. In this experiment, RpS3+/+ Xrp1+/+ cells were distinguished by brighter labeling for ß –galactosidase than for the RpS3+/− Xrp1+/− cells (darker magenta). U) Enlargement of the beta-galactosidase channel from panel T. Note small clones of RpS3−/− Xrp1−/− cells lacking beta-galactosidase. Supplemental data related to this Figure is shown in Figures S1 and S2, and in Table S1.
The effects of Xrp1 mutations on cell competition are illustrated in Figure 1. Normally, cell competition is expected in mosaic imaginal discs that contain both wild type and Rp+/− cells. The Rp+/− cells undergo competitive apoptosis near the wild type cells, which grow to predominate in the tissue, as illustrated when clones of RpS3+/+ cells grow in RpS3+/− wing imaginal discs (Figure 1B, D, E))(Morata and Ripoll, 1975; Li and Baker, 2007; Martin et al., 2009). We did not observe cell competition when an Xrp1 mutation was also present, however. Clones of Xrp1−/− cells growing in an RpS3+/− Xrp1+ wing disc induced very little competitive cell death and the Xrp1−/− clones occupied significantly less territory than wild type clones achieved in RpS3+/− imaginal discs (Figure 1C-E).
In another cell competition assay, where the imaginal discs are initially wild type and mitotic recombination generates clones of Rp+’ cells, cell competition would normally prevent such cells from growing and surviving for long(Morata and Ripoll, 1975; Simpson, 1979; Moreno et al., 2002; Tyler et al., 2007)(Figure 1F). In contrast to this, clones of RpL36+/−; Xrp1−/− cells survived and differentiated in RpL36+/+; Xrp1+/− animals (Figure 1G). Thus, Xrp1 mutation prevented cell competition between wild type and RpS3+/− or RpL36+/− genotypes.
To address how Xrp1 affects cell competition, we first looked at gene expression changes that have been reported during cell competition(Moreno et al., 2002; Portela et al., 2010). Previously, the extracellular matrix protein Sparc was reportedly induced in competing cells. In our hands the antibody mAb30A1B4 labeled a nuclear antigen expressed by all Rp+/− disc cells, regardless of proximity to Rp+/+ cells (Figure 1H-I). Whether or not this mAb30A1B4 staining in Rp+/− cells represents Sparc protein, it was eliminated by the Xrp1 mutation (Figure 1J-K). Likewise, and as also reported by others (Kucinski et al., 2017), we also found that Puc-LacZ was elevated in all Rp+/− cells, not only those close to wild type cells in mosaics (Figure 1L-M). The Xrp1 mutation reduced puc-LacZ expression in Rp+/− cells (Figure 1N-O).
Because Xrp1 affected gene expression that occurred throughout Rp+/− tissues, not just in Rp+/− cells in close proximity to wild type cells, we wondered whether Xrp1 might act directly in Rp+/− cells. In the experiments described above, Rp+/− cells were heterozygous for the Xrp1m2-73 mutation, suggesting this mutation acted dominantly. Consistent with this, no competition was observed between wild type and RpS18+/− cells in a Xrp1m2-73/+ background (Figures 1P-Q, S1A-D). The dominant function occurred cell-autonomously in the Rp+/− cells, because competition was also absent between wild type clones, lacking any Xrp1 mutation, and Xrp1m2-73/+ RpS3−/+ cells (Figures 1R-U, S1E).
The truncated proteins encoded by Xrp1m2-73 should lack both the AT hook and bZIP DNA binding domains and might lack Xrp1 function. To test whether loss of one Xrp1 gene copy affected cell competition, effects of chromosomal deletions were examined. Consistent with dominant haploinsufficiency, cell competition did not occur between wild type and RpS18+/− cells when one Xrp1 copy was mutated (Figure S1G-K). This was tested with Df(3R)Exel6181 and Df(3R)Exel6182 (Figure S1G-K), chromosome deletions that extend into the left or right parts of the Xrp1 locus, respectively (Figure 1A). By contrast, cell competition continued in heterozygotes for Df(3R)Exel6187, a similar deficiency that deletes other chromosomal regions but not the Xrp1 gene (Figure S1F). Cell competition was also lacking in the genetic background Xrp1D1/+ (Figure S1L,M). We generated Xrp1D1 through a targeted deletion by recombination between FRT sequences of known locations that deleted all of the Xrp1 locus and parts of the neighboring genes (Figure 1A). All these heterozygous genotypes reduced cell competition to a degree similar to genotypes that completely lacked any functional Xrp1 gene, including Xrp1m2-73/D1, Xrp1m2-73/Df(3R)Exel6182, and Df(3R)Exel6181/Df(3R)Exel6182, (Figure S1N-P). All these Xrp1 genotypes were viable and fertile as adults, indicating that Xrp1 is dispensable for wild type development and viability.
In the course of the experiments it became clear that removing Xrp1 function was able to ameliorate the effects even of homozygous Rp mutations. Most Rp mutations are cell-autonomously lethal as homozygotes, reflecting the essential role of most Rp in ribosome function(Lambertsson, 1998; Marygold et al., 2007). Mitotic recombination in the Rp+/− genotypes used in this paper generates Rp+/− cells as reciprocal recombinants to the Rp+/+ cells, but whereas Rp+/+ cell clones grow and outcompete Rp+/− neighbors, clones of Rp+/− cells are normally lost from imaginal discs soon after generation (Morata and Ripoll, 1975; Kale et al., 2015a)(Figure 1R,S). By contrast, clones of Xrp1−/− RpS3−/− cells survived in wing imaginal discs 72-96h h after induction of mitotic recombination, although they underwent only limited proliferation to attain a small size(Figure 1T-U). Sequencing the mutant RpS3 transcription unit revealed four nucleotide substitutions compared to the reference sequence (Figure S2). One was intronic, two were predicted to be silent but one mutates the ATG start codon to GTG, with no alternative ATG in frame from which RpS3 translation could be initiated (Figure S2). As with RpS3, small clones of RpS18−/− Xrp1m2-73/D1 cells were also observed in wing imaginal discs 72-96h h after induction of mitotic recombination (Figure S1N), and the same for RpS18−/− Xrp1m2-73/Df(3R)Exel6182 (Figure S1O), and RpS18−/− Df(3R)Exel6181/Df(3R)Exel6182 (Figure S1P). We don’t know if these cells survive indefinitely, however, since some were undergoing cell death (Figure S1N-P).
To determine whether Xrp1 was required for cell death in general, eye development was compared between wild type and the Df(3R)Exel6181/Df(3R)Exel6182 genotype that lacks a normal Xrp1 gene (Figure 2). During normal development, apoptosis eliminates supernumerary cells first from the pupal retina and then from the margins of the eye field(Wolff and Ready, 1991)(Figure 2A,C). Neither wave of apoptosis was affected in Df(3R)Exel6181/Df(3R)Exel6182, and the subsequent cellular composition of the retina was indistinguishable from wild type, confirming that developmental apoptosis in Df(3R)Exel6181/Df(3R)Exel6182 and wild type was quantitatively equivalent (Figure 2 B,D,E). These findings illustrated that Xrp1 function was not required for all developmental cell death, although it was required for cell death during cell competition.
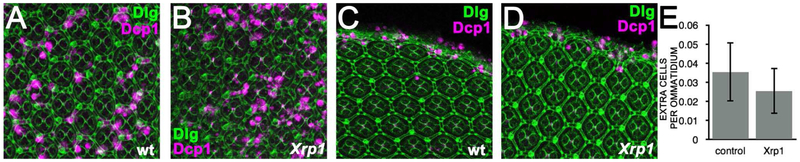
Figure 2. Xrp1 does not affect developmental cell death in the retina.
Panels A-D show pupal retinas labeled for Dlg protein to outline cells (green) and anti-active Dcp1 to reveal apoptotic cell death (magenta). A) 30h after puparium formation (APF). At this stage, apoptosis culls extra cells from the wild type retina. B) Similar cell death occurs in Xrp1−/− mutant retina 30h APF. C) At 43h APF, apoptosis removes cells from the retinal periphery. D) Cell death occurs similarly in Xrp1 mutant retina 43 APF. E) Supernumerary cells were counted 42 h APF to evaluate the efficiency of programmed cell death in the retina. There was no significant increase in supernumerary cells in the Xrp1−/− mutant, indicating that this developmental cell death occurred normally (t-test, p>0.05). Error bars indicate S.E.M. Supplemental data related to this Figure is shown in Table S1.
Xrp1 is a negative growth regulator
Our results suggested that Xrp1 could affect the growth of Rp+/’cells, as well as their competitiveness. This was because in addition to preventing competitive apoptosis, Xrp1 mutations also decreased the relative size of wild type clones compared to the Rp+/− Xrp1+/− areas of mosaic discs (Figure 1C,E, Q,T, Figure S1B,D, G, H, J). This could be explained if Rp+/− cells that are also mutated for Xrp1 grow more rapidly than plain Rp+/− cells, reducing the growth differential with Rp+/+ cells(Martin et al., 2009). To test this, the relative growth of RpS18+/− cells was compared with that of RpS18+/−; Xrp1−/− cells. These were obtained as reciprocal mitotic recombinants in RpS18+/−; Xrp1+/− imaginal discs (Figure 3A,B). The RpS18++/− ; Xrp1−/− clones grew at more than twice the rate of RpS18+/− Xrp1++/+ clones, indicating that wild type Xrp1 function retards the growth of RpS18+/− cells. This was specific to Rp+/− mutant cells, because Xrp1−/− clones grew identically to Xrp1+/+ controls in tissues that lacked any Rp mutation(Figure 3A-C).
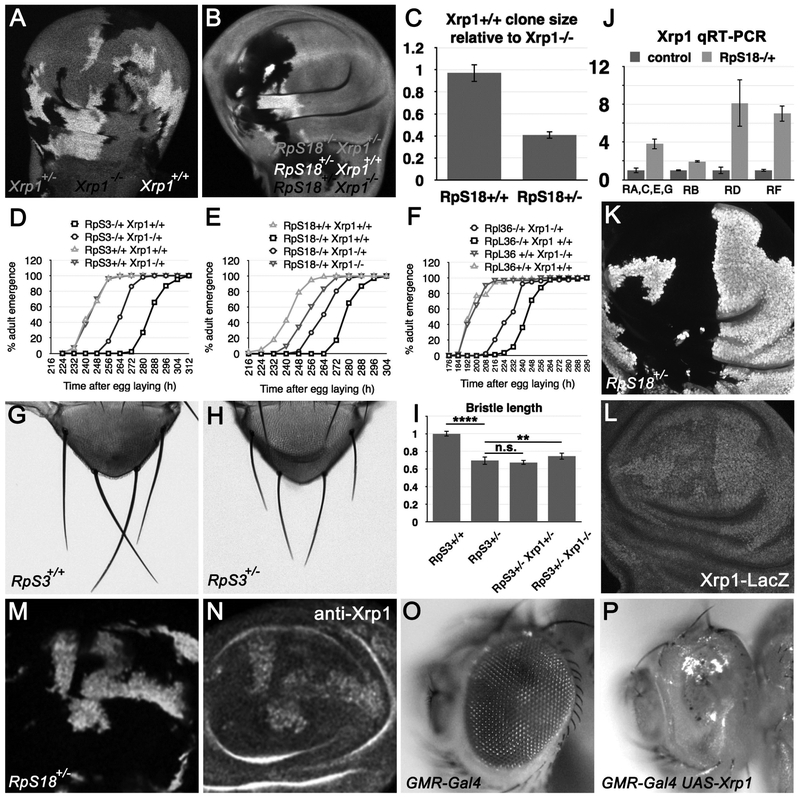
Figure 3. Xrp1 expression retards growth.
A) Xrp1+/− wing discs containing clones of Xrp1−/− and Xrp1+/+ cells. B) RpS18+/− Xrp1+/− wing discs containing clones of RpS18+/− Xrp1−/− and RpS18+/− Xrp1+/+ cells. C) Quantification of clones size data from experiments illustrated in panels A,B. D-F) Graphs representing developmental rate as time in hours from egg laying to adult emergence for the genotypes indicated. In panel D, the emergence times for RpS3+/+ Xrp1+/+ and RpS3+/+ Xrp1+/− were not significantly different (two-tailed Mann-Whitney test, p=0.542). In panel F, the emergence times for RpL36+/+ Xrp1+/+ and RpL36+/+ Xrp1+/− were not significantly different (two-tailed Mann-Whitney test, p=0.43). In panels D-F, the emergence of RpS3++/−, RpS18+/−, and RpL36+/− was each delayed significantly compared to the wild type controls (one-tailed Mann-Whitney test, p<0.00001 in all cases). The rescue of the respective Rp+/− Xrp1+/− adults was highly significant (one-tailed Mann-Whitney test, p<0.00001 in all cases). In panel E, the accelerated development of RpS18+/− Xrp1−/− compared to RpS18+/− Xrp1+/− was highly significant (significant (one-tailed Mann-Whitney test, p<0.00001) G) Scutellum of RpS3+/+ fly, showing the scutellar bristles. H) Scutellum of RpS3+/− fly, showing the scutellar bristles. See Figure S2 for more genotypes. I) Mean length of posterior scutellar bristles from genotypes indicated. RpS3+/− bristles were ~30% shorter than controls (p<0.0001, t-test). RpS3+/− Xrp1+/− bristles were no different. RpS3+/− Xrp1−/− bristles were slightly longer (~5%) and this difference was statistically significant (p<0.01, t-test). J) qRT-PCR measurements of Xrp1 transcript isoforms (see Figure 1A for diagram of isoforms). All the Xrp1 transcripts were elevated in RpS18+/− wing imaginal discs in comparison to wild type controls. See Figure S2 for qRT-PCR data compared to tubulin mRNA levels. K) Mosaic wing disc containing RpS18+/− cells (labeled for ubi-GFP) and RpS18+/+ cells (unlabeled). L) Xrp1-LacZ labeling of the wing imaginal disc shown in panel K. Since the LacZ mRNA encoded by the enhancer trap insertion contains no sequences from Xrp1, elevated LacZ labeling likely reflects enhanced transcription. M) Mosaic wing disc containing RpS18+/− cells (labeled for ubi-GFP) and RpS18+/+ cells (unlabeled). N) Anti-Xrp1 labeling of the wing imaginal dsc shown in panel M. O) Flies heterozygous for the GMR-Gal4 transgene had normal eyes. P) Ectopic Xrp1 expression under control of the GMR-Gal4 driver ablated the eye almost completely(Tsurui-Nishimura et al., 2013). Supplemental data related to this Figure is shown in Figure S3 and Table S1.
These results prompted us to examine the effect of Xrp1 on the overall growth of the organism. Whereas adults of the genotype RpS3+/− emerged 1.7 days later than controls, on average, heterozygosity for Xrp1 reduced the delay to 0.8 days, on average (Figure 3D). Similarly, whereas adults of the genotype RpS18+/− emerged 1.4 days later than controls, on average, the delay was reduced to 0.8 day by heterozygosity for Xrp1, and to 0.4 day in RpS18+/−Xrp1−/− (Figure 3E). These findings extended to mutations affecting the large ribosomal subunit. Adults of the genotype RpL36+/− emerged ~2 days later than controls, on average, the delay was reduced to ~1.3 days by heterozygosity for the Xrp1 (Figure 3F). In contrast to these effects on the rate of development, the size of bristles from RpS18+/− adults was much less affected by Xrp1 genotype (Figure 3G-I and Figure S3A-D).
Xrp1 expression is induced in Rp mutant cells to slow growth
In order to understand how Xrp1 might affect specifically the growth of Rp+/− genotypes, qRT-PCR was performed using RNA extracted from wing imaginal discs to examine Xrp1 transcription. In comparison to tubulin mRNA, the various isoforms of transcript were from 2-fold to 6-fold more abundant in RpS18+/− tissue (Figure 3J and Figure S3E). Transcription was also assessed independently through an enhancer trap P{PZ}Xrp102515, (Figure 1A). Little expression of this transcriptional reporter was detected in wild type imaginal discs but expression was elevated cell-autonomously in Rp+/− cells (Figure 3K-L). As would be expected from the elevated transcription, we also found using an antibody that Xrp1 protein levels were elevated in Rp+/− cells, compared to wild type (Figure 3M-N).
When over-expressed at high levels, Xrp1 acts as a potent inhibitor of cell proliferation in cultured cells and prevents growth and survival of imaginal disc cells (Akdemir et al., 2007; Tsurui-Nishimura et al., 2013). Transcriptional induction in the developing Drosophila eye disc using the Gal4 system ablates the adult eye almost completely (Figure 3O-P). These findings suggest that elevated Xrp1 transcription and protein, such as occur in Rp+/− cells, could be sufficient to restrict growth.
Xrp1 inhibits translation in Rp mutant cells
When cell cycle markers were compared between clones of Rp+/− Xrp1−/− cells and clones of Rp+/− Xrp1+/+ cells that were growing at different rates, no differences were seen in Histone H3 phosphorylation or Cyclin B expression, markers of progression through M-phase and S-G2-phases of the cell cycle respectively (Figure S4A-D and S4I-K). There was also no difference in average cell size (Figure S4E-G), which is consistent with prior studies reporting that RpS3+/− cells are the same size as wild type cells(Neufeld et al., 1998). These findings indicate that physiological levels of Xrp1 slowed the growth rate of imaginal disc cells without a major effect on one particular cell cycle transition.
To understand how growth was altered without any specific cell cycle transition, we investigated the rate of translation in imaginal discs. An alkyne analog of puromycin, O-propargyl-puromycin (OPP) has been used as a Click chemistry reagent for fluorescent labeling of nascent protein synthesis in tissue culture cells (Liu et al., 2012). When imaginal discs explanted from wild type larvae were incubated with OPP, incorporation was seen in somewhat patchy patterns (Figure 4A-B and Figure S4L-N). Although the inhomogeneity was unexpected, because proliferation is reportedly uniform at these wing disc stages, the translation patterns closely resemble those recently described for phospho-RpS6 and which reflect TORC1-dependent growth in the wing imaginal disc(Romero-Pozuelo et al., 2017). OPP incorporation was abolished by cycloheximide, indicating dependence on translation (Figure 4E), and reduced by knockdown or mutation of genes that promote translation, including Tor, myc, and GADD34, a phosphatase that dephosphorylates eIF2a (Figure 4C-D and Figure S4O-T). Taken together with the correlation with TORC1 signaling, these controls justify use of OPP incorporation to measure bulk translation rates in imaginal discs.
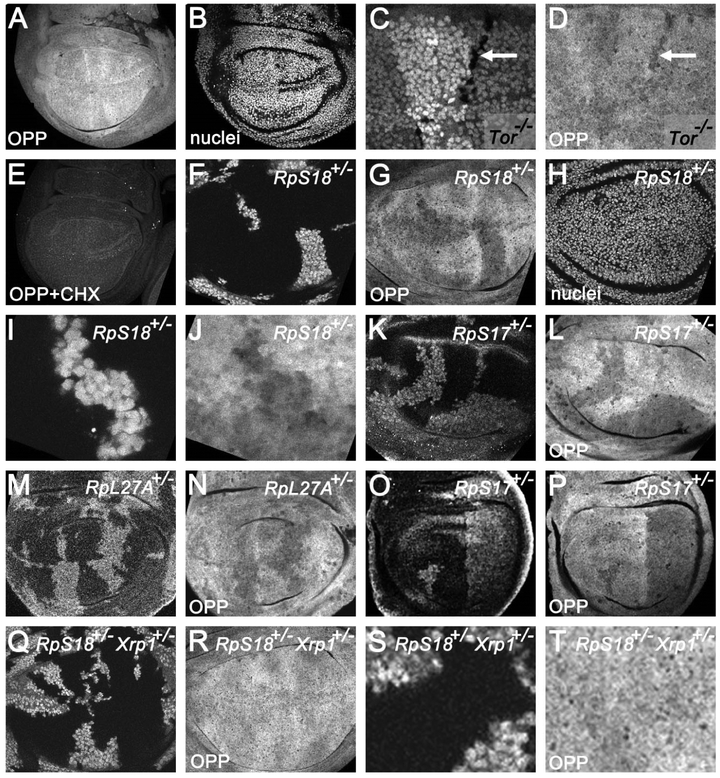
Figure 4. Protein synthesis in wing imaginal discs.
A) Incorporation of o-propargyl puromycin (OPP) revealed mostly similar translation rates across the wing disc although with some patchiness. A single confocal plane is shown here. These patchy patterns seemed similar although not identical in individual wing discs (see Figure S3L-N for more examples). B) Since most newly-synthesized protein is nuclear, it is important to note that fluctuations in translation rate did not simply correlate with presence of nuclei in the confocal plane. C) Enlarged portion of wing disc containing dTor−/− clones (unlabelled cells, arrow). Only small dTor−/− clones were recovered, reflecting reduced growth. D) dTor−/− incorporated less OPP (arrow). E) OPP incorporation was inhibited by cycloheximide, an inhibitor of translation. F) Mosaic wing disc containing RpS18+/− cells (labeled for GFP) and RpS18+/+ cells (unlabeled). G) Reduced OPP labeling of RpS18+/− cells. H) Nuclei throughout the affected focal planes. I) Enlarged region from panel F containing RpS18+/− cells. J) OPP incorporation in the enlarged region from panel I. K) Mosaic wing disc containing RpS17+/− cells (labeled for GFP) and RpS17+/+ cells (unlabeled). L) Reduced OPP labeling of RpS17+/~ cells. M) Mosaic wing disc containing RpL27A+/’ cells (labeled for beta-galactosidase) and RpL27A++ cells (unlabeled). N) Reduced OPP labeling of RpL27A+/’ cells. O) Mosaic wing disc containing RpS17+/− cells (labeled for GFP). In this example RpS17+/+ cells (unlabeled) were seen only in the anterior compartment, which they had colonized almost completely. P) Reduced OPP labeling of RpS17+/− cells. Cells of the posterior compartment, which were not in competition with RpS17+/+ cells, showed translation rates as low as those RpS17+/− cells remaining in the anterior compartment that were competing with RpS17+/+ cells. Q) Mosaic wing disc from an Xrp1+/− larva containing RpS18+/− Xrp1+/− cells (labeled for GFP) and RpS18+/+ Xrp1+/− cells (unlabeled). R) OPP incorporation in the two genotypes was indistinguishable. S) Enlargement of the wing disc from panel U. T) OPP incorporation in the enlarged portion of the wing disc from panels U, V. Supplemental data related to this Figure is shown in Figure S4 and Table S1.
When translation rates were compared between wild type and Rp+/− cells in the same imaginal discs, lower translation was consistently seen in Rp heterozygous cells (RpS18+/−, RpS17+/−, RpL27A+/− and RpS3+/− were examined)(Figure 4F-N and data not shown). To assess whether lower translation was a cell-autonomous property of Rp+/− cells, or might be due to competition with wild type cells, we examined discs where A and P compartments were of different genotypes, since cells do not compete across the compartment boundary(Simpson and Morata, 1981). These experiments showed that translation rate was cell-autonomously lower in Rp+/− compartments than in wild type cells in the other compartment, indicating that it was a property of Rp+/− cells not experiencing cell competition (Figure 4O-P).
When RpS18+/− cells were studied in the presence of Xrp1 mutations, the difference in translation rate between wild type and RpS18+/− cells could no longer be detected (Figure 4Q-T). Taken together, these results suggest that RpS18+/− cells genotypes grow more slowly as a result of reduced translation rate, and that Xrp1 is responsible for the reduced translation rate.
Xrp1 has little effect on ribosome number
One factor that could affect translation rates was the number of ribosomes. To assess how mutations in Rp genes or Xrp1 affected ribosome biogenesis, we used northern blotting to estimate the steady state levels of mature rRNA in wing imaginal disc cells. In Drosophila as in other eukaryotes, 28S, 18S and 5.8S rRNA are processed from a common precursor RNA during the assembly of the ribosomal subunits(Long and Dawid, 1980; Kressler et al., 2017). For the SSU, a final cleavage generates the 18S rRNA from a 20S species once assembled SSU are exported into the cytoplasm(Ferreira-Cerca et al., 2005). For the LSU, trimming of the 28S and 5.8S rRNA’s to their mature forms may be a requirement for the nuclear export machinery(Ohmayer et al., 2013). Thus levels of these mature rRNA’s are expected to reflect numbers of mature ribosomal subunits.
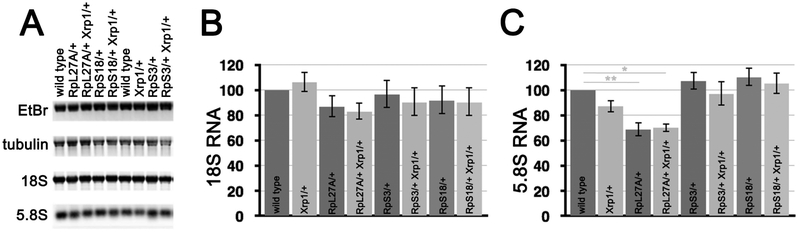
When equal levels of total RNA were blotted from wild type, RpS18+/− wing discs, RpS3+/− wing discs, and RpL27A+/− wing discs, similar levels of 18S rRNA were detected, when normalized against actin or tubulin mRNAs in the same samples, indicating similar concentrations of mature SSU in these genotypes relative to mRNA (Figure 5A,B). However, lower levels of 5.8S rRNA were detected in RpL27A+/− wing discs, consistent with reduced levels of LSU in this RpL mutation only, not in the two RpS mutants (see Figure 5 legend for statistical comparisons of rRNA levels between genotypes). It is most convenient to monitor LSU through the 5.8S rRNA band (Figure 5C) because the 28S rRNA is further cleaved in Drosophila to forms that run on top of the 18S rRNA.
Figure 5. Ribosome levels in wing imaginal discs.
A) Northern blotting of total RNA samples from wing imaginal discs with multiple probes. Ethidium bromide staining verifies gel loading and membrane transfer of similar total RNA amounts from samples. The majority of total RNA is rRNA, and since the mature 28S rRNA is processed in Drosophila into two similarly-sized fragments, the 18S rRNA and 28S rRNA products appear as a single overlapping band. 18S rRNA (a component of mature SSU) and 5.8S rRNA (a component of mature LSU) were detected using specific hybridization probes. The rRNA levels were normalized against mRNA for tubulin or actin detected on the same blots. mRNA-Seq has indicated that tubulin and actin mRNA levels are similar in wild type and RpS3/+ wing discs)(Kucinski et al., 2017 and data not shown), assuming that overall mRNA concentrations are similar between wild type and Rp+/− wing discs. B) 18S rRNA level in indicated genotypes in comparison to wild type. Geometric means from 4-6 biological replicates are shown. Error bars represent ± 1 S.E.M. of the log-transformed data. No significant differences are observed among the genotypes (1-way ANOVA, p=0.6824), although there is a trend for 18S levels to be somewhat reduced in all the Rp+/− genotypes. In addition, in no case were genotypes with and without the Xrp1 mutation significantly different (p>0.05 for paired t-tests in all cases). C) Analysis of 5.8S rRNA performed as in panel B. 5-6 biological replicates are shown. The null hypothesis that 5.8S levels are the same in all samples is rejected (1-way ANOVA, p=0.000011). Pairwise comparison of mutant genotypes to wild type control indicates significant differences only between wild type and RpL27A/+ (Bonferroni-Holm method, p<0.01) and between wild type and RpL27A/+ Xrp1/+ (p<0.05). In addition, in no case were genotypes with and without the Xrp1 mutation significantly different (p>0.05 for paired t-tests in all cases). Supplemental data related to this Figure is shown in Tables S1-S3.
These same analyses were also performed using wing discs from the same genotypes but also heterozygous for the Xrp1 mutation, for which restoration of growth and/or translation rates had been observed. In no case did Xrp1 mutation much affect the relative ribosome content. That is, steady-state rRNA levels (relative to mRNA) were not significantly different between RpS18+/− and RpS18+/− Xrp1+/− wing discs, RpS3+/− and RpS3+/− Xrp1+/− wing discs, or between RpL27A+/− and RpL27A+/− Xrp1+/− wing discs (Figure 5B-C). We also noted similar rRNA levels in wild type and Xrp1+/− wing discs (Figure 5B-C).
Taken together, these findings suggest that wing discs from three Rp+/− genotypes might contain similar SSU subunit levels to wild type wing discs. LSU levels were also similar in two mutants where the affected Rp was a component of the SSU, but a mutation in the LSU protein RpL27A reduced LSU numbers. If RpS3 and RpS18 mutations reduced translation without much reducing levels of ribosomal subunits, and Xrp1 had little effect on subunit numbers, either in wild type or in any of the Rp+/− genotypes, then Xrp1 expression may reduce translation rate of Rp+/− genotypes through some other mechanism.
Xrp1 regulates transcriptional responses in Rp/+ wing discs
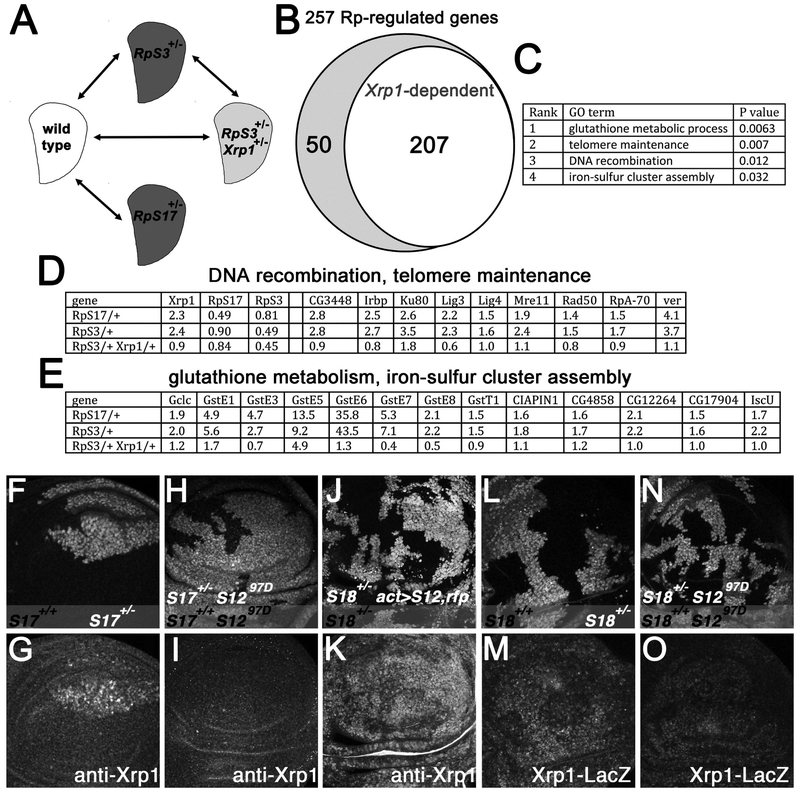
Since Xrp1 is even responsible for the changes in overall translation in Rp+/− cells, we wondered whether Xrp1 controls all the effects of Rp+/− mutations. Hundreds of genes show altered transcription in Rp+/− wing discs, including components of the DNA damage response, oxidative stress response, innate immune response, Jnk pathway and Jak/Stat pathway(Kucinski et al., 2017). To assess how these transcriptional effects depended on Xrp1, we first isolated mRNA from wing imaginal discs from two independent genotypes, RpS3+/− and RpS17+/− for analysis by mRNA-Seq (Figure 6A). In comparison to parallel wild type controls, transcripts of 253 genes (189 increased and 64 decreased) were significantly altered in both Rp+/− genotypes, representing a shared response common to multiple Rp+/− genotypes (Figure 6B; Table S2). These genes were enriched for GO terms related to DNA repair and to oxidative stress(Figure 6C-E; Figure S4A). To validate the mRNA-Seq data, qRT-PCR was performed for one representative gene from each of the enriched GO-term classes (Irbp18, Mre11, GstE6, and Iscu), and each showed a significant change commensurate with the mRNA-Seq data (Figure S4B). These mRNA-Seq results verified the elevation of Xrp1 transcripts in both RpS17+/−, and RpS3+/−, and also documented 50% reduction in mRNA levels for RpS3 in RpS3+/−, and for RpS17 in RpS17+/−, which confirms that reduced expression of the ribosomal protein genes is the primary defect in these genotypes (Figure 6D).
Figure 6. Xrp1 regulates many genes and is induced by RpS12.
A) mRNA was isolated from third instar wing discs enabling comparisons between four genotypes. B) Transcript levels of 253 genes were altered in both RpS17++/− and RpS3++/− and 206 of these changes were regulated by Xrp1. C) 4 GO terms for Biological Process were significantly enriched in the Rp+/− -regulated genes (ie P<0.05 after Benjamini correction). D) Expression levels (fold changes relative to wild type control according to Deseq2) for Xrp1, RpS17, RpS3 and for the 9 Rp+/−-regulated genes with telomere maintenance and DNA recombination GO terms, all of which were regulated by Xrp1. E) Expression levels (fold changes relative to wild type control according to Deseq2) for the 13 Rp+/−-regulated genes with glutathione metabolic process and iron-sulfur cluster GO terms, all of which were regulated by Xrp1. F) RpS17+/− wing imaginal disc almost entirely occupied by RpS17+/+ clones (unlabelled cells). G) Elevated Xrp1 protein levels in RpS17+/− cells. H) RpS17+/− rpS1297D/97D wing imaginal disc almost entirely occupied by RpS17+/+ rpS1297D/97D clones (unlabelled cells). I) Xrp1 protein is not elevated in the RpS17+/− rpS1297D/97D cells. J) Clonal expression of RpS12 and RFP proteins RpS18+/− wing disc. K) Xrp1 protein is elevated by RpS12 over-expression. L) RpS18+/+ clones (unlabelled cells) in RpS18+/− wing disc. M) Xrp1-LacZ is elevated in RpS18+/− cells. N) RpS18++/+ rpS1297D/97D clones (unlabelled cells) in RpS18+/− rpS1297D/97D wing disc. O) Xrp1-LacZ is not strongly elevated in the RpS18+/− rpS1297D/97D cells. Supplemental data related to this Figure is shown in Figure S5 and Table S1.
Since there is evidence that cell competition may share components with innate immune signaling (Meyer et al., 2014), it is worth mentioning that several genes related to innate immunity were elevated in Rp+/− wing discs although such genes were not statistically enriched (Figure S5A). Other genes implicated in oxidation-reduction process, Jak-Stat signaling, and JnK signaling were among those altered in both RpS17+/−, and RpS3+/− wing discs, although these GO terms were not statistically enriched by our criteria (Figure S5A).
When mRNA was isolated from RpS3+/− Xrp1+/− wing discs, we found that transcription of RpS3 was not restored (Figure 6D). Although there was also a modest (10-20%) reduction in RpS17 transcription in RpS3+/− mutants and vice versa, RpS17 transcription was not restored in RpS3+/− Xrp1+/− wing discs either (Figure 6D). Thus there was no evidence for any role of Xrp1 in ribosome biogenesis through transcription of Rp genes. By contrast, we found that >81 % of the protein gene expression changes common to Rp+/− genotypes depended on Xrp1 (206 genes) (Figure 6B). This may represent an underestimate of the dependence on Xrp1, since perhaps some genes would only be affected by homozygous mutation of Xrp1. The Xrp1-dependent genes were enriched for the same biological process GO-terms as the complete set of Rp+/−-regulated genes, and included all 4 genes validated by qRT-PCR, which further supported Xrp1-dependent transcription for these genes (Figure S4B). Out of the 253 Rp+/−-regulated genes, 159 were also identified in a previous study of RpS3+/− wing discs(Kucinski et al., 2017)(Table S3). We found that 142 of these 159 genes, or >89%, were regulated by Xrp1. Taken together, these results demonstrated that, at least with regard to transcription, the large majority of changes in Rp+/− wing discs were mediated by Xrp1.
Xrp1 responds to an RpS12-dependent ribosomal distress signal
The finding that Xrp1 seems to be responsible for, and therefore upstream of the bulk translation defects of Rp+/− cells, raised the question of what mechanism upregulates Xrp1 transcription in Rp+/− genotypes. The Xrp1 gene was first recognized as a gene transcriptionally induced by p53 in response to irradiation(Brodsky et al., 2004). However, p53 is dispensable for competition of Rp+/− cells by wild type, suggesting that p53 is unlikely to induce Xrp1 expression in Rp+/− cells(Kale et al., 2015a). In agreement with this, dominant-negative p53 did not affect the expression of the enhancer trap P{PZ}Xrp102515 in Rp+/−, indicating that Xrp1 transcription is elevated in Rp+/− cells independently of p53 (Figure S5C-F).
Recently, we reported that a specific ribosomal protein, RpS12, plays a special role in cell competition, different from other ribosomal proteins(Kale et al., 2018). A missense mutant allele, rpS12G97D, protects Rp+/− genotypes from competition with wild type cells, whereas the wild type rpS12 allele is required for cell competition to occur. Accordingly, cells heterozygous for mutations in other Rp genes are not eliminated by competition with wild type cells if they also carry the rpS12G97D allele in place of the rpS12+ allele, whereas extra copies of the wild type rpS12 gene, or overexpression of the RpS12 protein, render cells heterozygous for other Rp mutations even less competitive(Kale et al., 2018). We therefore wondered whether rpS12, rather than p53, was required for transcription of Xrp1 in Rp+/− cells. We found that an rpS12 allele that prevents cell competition also blocked induction of Xrp1 protein in Rp+/− wing discs (Figure 6F-I). Conversely, when RpS12 was overexpressed in Rp+/− wing discs, Xrp1 expression was further elevated (Figure 6J-K). Since expression of the Xrp1-LacZ enhancer trap in Rp+/− wing discs was also RpS12-dependent, an RpS12 activity directly or indirectly induces Xrp1 transcription in Rp+/− cells (Figure 6L-O).
DISCUSSION
Most ribosomal protein genes are essential. Reflecting this importance, ribosomal protein mutations have dominant effects through haploinsufficiency. We have identified a bZip-domain protein, Xrp1, which behaved like a master-regulator of responses to Rp mutations (Figure 7). Even the acute lethality of Rp+/− cells depended on Xrp1. The only aspect of the Rp+/− phenotype that appeared largely independent of Xpr1 was the reduced size of the bristles, which was only slightly restored by Xrp1 mutations (Figure 3I and Figure S2A-D). Bristle size might depend on ribosome function directly, or on a different regulatory gene that replaces Xrp1 in bristle precursors.
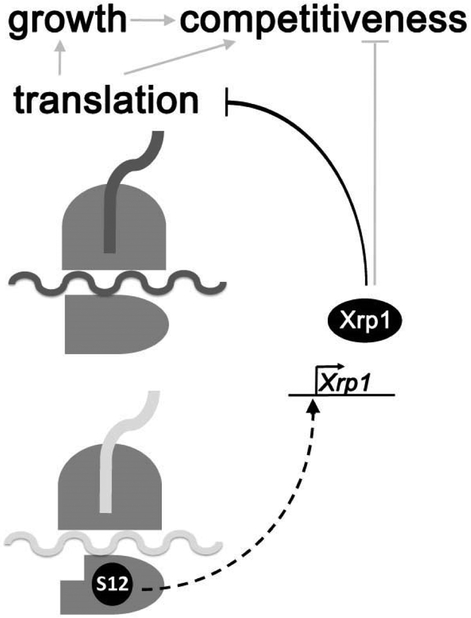
Figure 7. Model.
Rp/+ genotypes express a phenotype of reduced translation, slow cellular growth rate, and reduced competitiveness in comparison to wild type cells. All of these effects of the Rp+/− genotypes depended on the bZip-domain protein Xrp1. The signal to induce Xrp1 transcription depended on the RpS12 protein, which appeared to signal the existence of a ribosomal defect or ribosomal protein imbalance(Kale et al., 2018). Xrp1 was responsible for reducing the bulk translation rate in Rp+/− cells, and this must include a reduction in the translational activity of mature ribosomes. Reduced translation was likely responsible for the slow growth of Rp+/− cells, although Xrp1 might also affect growth independently of translation. Xrp1 also controlled the competitiveness of Rp+/− cells in mosaics with wild type cells, which provides a possibility for eliminating Rp+/− cells in favor of non-mutant replacements. We hypothesize that one or more target genes control competitiveness, either in response to Xrp1 itself, or indirectly in response to the changes in translation or growth rates. By utilizing cell competition, a decision could be made to eliminate defective cells only where better cells were available to replace them.
We first isolated a null allele of Xrp1 in a screen for mutations preventing cell competition. We found that Xrp1 transcription and Xrp1 protein were selectively elevated in Rp+/− cells and required cell-autonomously to render these cells less competitive than wild type cells (Figure 1 and Figure S1). Later we found that Xrp1 also acted to reduce the growth rate of Rp+/− cells (Figure 3A-C). In the absence of Xrp1, or even when Xrp1 gene dose was reduced to one copy, Rp+/− cells grew more like wild type cells. Xrp1 contributed substantially to the developmental delay of Rp+/− animals, which without Xrp1 could reach adulthood only slightly later than wild type animals, despite lacking one copy of essential Rp genes (Figure 3D-F).
Xrp1 probably reduces growth by reducing overall translation rate. Rp+/− cells had lower translation rates than wild type, but it was Xrp1, not haploinsufficiency for an important Rp gene, that reduced translation rate, because the difference from wild type disappeared when Xrp1 was mutated simultaneously (Figure 4 and Figure S4). Although ribosome numbers have not been counted directly, the proportion of rRNAs was not reduced in most Rp+/− genotypes, suggesting that an Xrp1-dependent reduction in translational activity per ribosome may occur. A mutation in RpL27A was the exception that did appear to reduce LSU number, but this was not rescued in RpL27A+/− Xrp1+/− discs and so was not responsible for the Xrp1-dependent growth inhibition (Figure 5). The persistence of Rp+/− mutant clones in the absence of Xrp1 also suggests changes in ribosome activity. Rp+/− cells should be deficient in synthesizing new ribosomes, so the prolonged survival of Rp+/− Xrp1−/− clones cannot easily be explained through restored ribosome biogenesis.
These findings suggest Xrp1 influences the rate of translation by cytoplasmic ribosomes, but they do not exclude additional roles in ribosome biogenesis. For example late third instar wing discs from wild type, RpS18+/−, and RpS18+/− Xrp1+/− larvae contained indistinguishable ribosome numbers (Figure 5), but these genotypes developed at different rates (Figure 3D-F). Based on the time taken for adults to emerge, we estimate that the ribosomes in late third instar wing discs had accumulated over ~80h, ~115h, and~100h of larval life respectively, so this is consistent with different rates of ribosome biogenesis generating similar absolute numbers of ribosomes over different durations of larval development. A reduced rate of ribosome biogenesis was reported previously in the mouse RpL24Bst/+ mutant (Oliver et al., 2004).
Regarding the overall rate of organismal development, it is well known that progress through the insect lifecycle is controlled in part through systemic signals ultimately controlling ecdysone levels(Boulan et al., 2015). This is not the primary means that Xrp1 affects imaginal disc growth because this occurs cell-autonomously (Figure 3A-C). Additional, non-autonomous effects of Xrp1 on organismal development are not ruled out. For example Dilp8, a secreted factor that regulates organismal growth(Boulan et al., 2015), undergoes Xrp1-dependent upregulation in Rp+/− wing discs(Figure S5A).
Hundreds of genes show altered mRNA levels in Rp+/− wing discs but it has not been clear how these changes arise (Kucinski et al., 2017). We now report that >80% of altered mRNA levels were Xrp1-dependent (Figure 6B). Some of these genes might be indirect targets of Xrp1, for example, Xrp1-dependent changes in overall translation rate may change gene transcription through a variety of mechanisms. The Xrp1-dependent changes include oxidative stress responses, which are reported to make cells less competitive (Kucinski et al., 2017), and also DNA repair genes (Figure 6C-E and Figure S5A).
How is Xrp1 induced by Rp mutations? Xrp1 is a transcriptional target of p53 in the response to irradiation, but p53 is not required for the elimination of Rp+/− cells by cell competition(Kale et al., 2015b). Accordingly, we showed that p53 was not required for Xrp1 induction in Rp+/− wing discs (Figure S5C-F). Reduced overall translation was unlikely to induce Xrp1 because Xrp1 was actually responsible for this. Perhaps more subtle changes in the translation of specific mRNA’s occur first and induce Xrp1 expression. Another possibility is that a signal is sent when ribosome assembly is altered, for example through an accumulated assembly intermediate. We report here that a particular ribosomal protein, RpS12, that was already recognized as a gene required for cell competition (Kale et al., 2018), was required to elevate Xrp1 transcription (Figure 6J,K). Although we don’t yet know the molecular mechanism by which RpS12 can affect transcription, this demonstrates that a link between a particular ribosomal protein and the Xrp1 gene triggers most of the response that occurs to mutations in other Rp genes, upstream of overall changes in bulk translation rates, which are a later consequence of the Xrp1 pathway. All the DNA repair gene expression in Rp+/− wing discs was also downstream of Xrp1 (Figure 6D), and may reflect Xrp1’s other role in the response to irradiation(Brodsky et al., 2004; Akdemir et al., 2007). How DNA repair genes contribute to aspects of the Rp+/− phenotype remains to be determined.
The importance of Xrp1 extends to homozygous Rp mutant cells. Remarkably, even Rp+/− cells survived and underwent limited growth if Xrp1 was completely removed. We don’t believe imaginal disc cells can grow and divide without ribosomes and protein synthesis, but since ribosome turnover occurs very slowly (Hirsch and Hiatt, 1966; Nikolov et al., 1983), Rp+/− recombinant cells probably retain most of the ribosome complement from the Rp+/− mother cell at first. They would be deficient in replenishing their ribosome complement, however, which would dilute with further growth and cell division. Previous studies indicate that when ribosome activity diminishes below a critical threshold, Rp+/− cells undergo apoptosis(Kale et al., 2015b). The absence of Xrp1 allowed Rp+/− cells to survive longer, by allowing more translation by the remaining ribosomes, and possibly by preventing competitive elimination of Rp+/− cells by Rp+/− cells.
Despite the unquestioned importance of ribosomal proteins in ribosome structure and function, our results indicate that the effects of Rp mutations in Drosophila are largely due to a regulatory response (Figure 7). Even the reduced translation in Rp mutant genotypes, which has also been observed in other organisms (Oliver et al., 2004; Cmejlova et al., 2006; Signer et al., 2014), is downstream of Xrp1 and does not play a primary role as a sensor of Rp mutations. Even when Rp genes are homozygously mutated, which seemingly should affect overall translation very quickly, Xrp1 normally kills the Rp+/− cells before such effects become evident. Rp+/− cells can also be killed by Xrp1, but indirectly, by cell competition when wild type cells are nearby.
Mutations that reduce translation or ribosome biogenesis by other routes are phenotypically distinct from Rp mutants. For example, mutations in the myc gene homolog or in components of the Drosophila TOR pathway lead to smaller flies, unlike Rp+/− flies (Gallant et al., 1996; Schreiber-Agus et al., 1997; Bohni et al., 1999; Montagne et al., 1999). In mammals also, mutations that affect ribosome biogenesis independently of ribosomal protein genes lead to human disease but the symptoms of such ribosomopathies differ from Diamond Blackfan Anemia (McCann and Baserga, 2013; Armistead and Triggs-Raine, 2014). These differences may occur because translation is affected indirectly in mutations of Rp genes and not the primary trigger for the cellular responses leading to pathology.
Although it may at first seem surprising that mutations in ribosomal proteins that are so directly involved in translation affect the cell through another mechanism, perhaps it is advantageous to mount such a coordinated response, for example to enable cell competition. Much as it is adaptive to eliminate cells with damaged DNA through apoptosis, perhaps cell competition is a mechanism to eliminate one or a few cells with defective ribosomes in favor of other, more normal cells.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nicholas Baker (Nicholas.baker@einstein.yu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experimental Animals
Species: Drosophila melanogaster. Strains were generally maintained at 25°C on medium containing the following ingredients per 1L: 18g yeast; 22g molasses; 80g malt extract; 9g agar; 65g cornmeal; 2.3g methyl para-benzoic acid; 6.35ml propionic acid. Experiments to measure the developmental rate of fly strains (Figure 4) used yeast-glucose media(Sullivan et al., 2000) or the semi-defined medium of Backhaus(Backhaus et al., 1984). Sex of larvae dissected for most imaginal disc studies was not differentiated.
METHOD DETAILS
Fly Stocks:
Previously existing genetic strains are described in the Key Resources Table: Xrp1… was generated as a Flp-mediated excision between P(XP)d11439 and PBac(WH)f07598 and is a deletion of sequences 3R:18,912,939-18,938,334, completely encompassing the Xrp1 gene.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Streptavidin IRDye 800 CW | LiCor (1:10000) | P/N 926-32230 |
| mouse anti-b-Galactosidase | DSHB (1:100) | JIE7 |
| mouse anti-b-Galactosidase (mAb40-1a) | DSHB (1:100) | ID: AB_2314509 |
| rat anti-GFP | Nacalai Tesque (1:500) | Cat# GF090R; RRID: AB_2314545 |
| rabbit anti-active-Dcp1 | Cell Signaling Technology (1:50) | Cat #9578 |
| mouse anti-discs large (4F3) | DSHB (1:50) | ID: AB_528203 |
| Mouse anti-dSparc | Portela et al., 2010 | mAb30A1B4 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Heat inactivated Fetal Bovine Serum | Gibco | 10082139 |
| Schneider's Drosophila Medium | Gibco | 21720024 |
| Trizol | Ambion | 15596-026 |
| Cycloheximide | Sigma | C1988 |
| Critical Commercial Assays | ||
| Click-iT® Plus OPP Alexa Fluor® 594 Protein Synthesis Assay Kit | Thermofisher | C10457 |
| Maxiscript T7 Transcription kit | Ambion | AM1312 |
| Deposited Data | ||
| mRNA-Seq raw data | This paper | GSE112864 |
| DESEQ2 analysis wildtype vs RpS17/+ | This paper | GSE112864 |
| DESEQ2 analysis wildtype vs RpS3/+ | This paper | GSE112864 |
| DESEQ2 analysis wildtype vs RpS3/+ Xrp1/+ | This paper | GSE112864 |
| DESEQ2 analysis RpS3/+ vs RpS3/+ Xrp1/+ | This paper | GSE112864 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster mutation Tor[DeltaP] | (Zhang et al., 2000) | FBal0120586 |
| D. melanogaster transgenic strain P{TRiP.JF01761}attP2 (mycRNAi) | (Ni et al., 2011) | FBti0114534 |
| D. melanogaster transgenic strain P{y[+t7.7] v[+t1.8]=TRiP.HMS00811}attP2 (Gadd34RNAi) | Bloomington (Ni et al., 2011) | FBti0140523 |
| D. melanogaster mutation Xrp1m2-73 | Lee et al., 2016 | N/A |
| D. melanogaster mutation Xrp1Δ1 | This paper | N/A |
| D. melanogaster mutation RpS3 | (Burke and Basler, 1996) | N/A |
| D. melanogaster transgenic strain UAS-Xrp1-Flg-CDS2 | Tsurui-Nishimura et al., 2013 | N/A |
| D. melanogaster transgenic strain P{PZ}Xrp1[02515] | Spradling et al., 1999 | Flybase: FBal0009448 |
| D. melanogaster transgenic strain P{GUS-p53.259H} | Brodsky et al., 2000 | Flybase: FBti0023695 |
| D. melanogaster transgenic strain P{GUS-p53.Ct} | Brodsky et al., 2000 | Flybase: FBti0023694 |
| D. melanogaster transgenic strain en-Gal4 | (Neufeld et al., 1998) | RRID:BDSC_6356 |
| D. melanogaster mutant strain Df(3R)Exel6181 | Exelixis, Inc. | Flybase: FBab0038236 |
| D. melanogaster mutant strain Df(3R)Exel6182 | Exelixis, Inc. | Flybase: FBab0038237 |
| D. melanogaster mutant strain Df(3R)Exel6187 | Exelixis, Inc. | Flybase: FBab0038242 |
| D. melanogaster strain Df(1)su(s)R194 (deleting RpL36 gene) | (Duffy et al., 1996) | FBab0024817 |
| D. melanogaster strain M(2)56f (mutating RpS18) | Laboratory of Y. Hiromi | FBal0284387 |
| D. melanogaster, RpS17 mutation M(3L)67C4 | Morata and Ripoll, 1975 | Flybase: FBal0011935 |
| D. melanogaster mutant for RpL27A Df(2L)M24F11 | Marygold et al., 2007 | Flybase: FBab0001492 |
| D. melanogaster. RpS12 mutation rpS12G97D | Tyler et al., 2007 | FBal0193403 |
| D. melanogaster UAS-rpS12 transgenic strain | (Kale et al., 2018) | FBal0337979 |
| D. melanogaster. RpL36+ transgenic strain | Tyler et al., 2007 | FBal0193398 |
| D. melanogaster. act>CD2>Gal4 expressing strain | Pignoni and Zipursky, 1997) | N/A |
| Oligonucleotides | ||
| See Table S4 | ||
| Software and Algorithms | ||
| ImageJ | (Schneider et al., 2012) | v1.44j |
| Photoshop CS5 | Adobe | V12.0.1 |
| Other | ||
| Biodyne B Nylon membrane | Thermofisher | 77016 |
| ULTRAhyb-Oligo buffer | Ambion | AM8663 |
| Odyssey Blocking buffer (PBS) | Li-COR | 927-40003 |
| Biotin-16-UTP | Roche | 11388908910 |
| RNA Sample Loading Buffer | Sigma | R4268-5VL |
Clonal analysis:
Genetic mosaics were generated using the FLP/FRT system employing hsFLP and eyFLP transgenic strains(Golic, 1991; Xu and Rubin, 1993; Newsome et al., 2000). For making clones using inducible hsFLP, larvae of non-Minute genotypes were subjected to 1 hour heat shock at 37°C, 60 ± 12 hours after egg laying and dissected 60hr later. For Minute/+ genotypes, heat shock was administered after 84 ± 12 hours of egg laying and dissected 72 hours later. Full genotypes for all figures are listed in Supplemental Table 1.
Measurement of developmental timing:
Development was monitored in cohorts of eggs laid within 8h periods. Cultures were monitored for adult emergence every 8h. Whenever possible experimental and control genotypes developed within the same vials and therefore same culture conditions, and were distinguished as adults using appropriate genetic markers. Where this could not be achieved, genotypes in common between the cultures were compared to assure comparable developmental rate.
Immunohistochemistry and Antibody labeling:
For antibody labeling, imaginal discs were dissected from late 3rd instar larvae in 0.1 M sodium phosphate buffer (pH 7.2) and fixed for 20 min at room temperature in 0.1 M PIPES (pH 6.95), 3.7% formalin, 2 mM MgSO4, 1 mM EGTA. Fixed imaginal discs were washed in PDT (0.3% sodium deoxycholate, 0.3% Triton X-100, 0.1 M sodium phosphate (pH 7.2) for 15 min on ice. Discs were incubated in primary antibody in PDT for overnight at 4°C, washed 3 times with PDT for 5 min each on ice and incubated in secondary antibody in PDT for 2-4 hours at 4°C, and washed 3 times with PDT for 5 min each on ice. After washes, discs were rinsed in 0.1 M sodium phosphate (pH 7.2) and incubated in glycerol mounting solution (75% (v/v) glycerol, 2% (m/v) n-propyl gallate) for 15 min on ice before mounting(Baker et al., 2014). Primary Antibodies are described in the Key Resources Table. Secondary Antibodies were Cy2-, Cy3- and Cy5-conjugates from Jackson Immunoresearch.
Image Acquisition and Processing.
Confocal images were recorded using Leica SP2 and SP5 confocal microscopes using 20× and 40× objectives. Images were processed using Image J1.44j and Adobe Photoshop CS5 Extended. Clone areas and perimeters were quantified from single confocal z-planes using the ‘Measure’ function in Image J 1.44j. Adult fly thoraces were dehydrated through an alcohol series and mounted in DPX medium for photography using a Zeiss Axioplan microscope. Thoracic bristle images were recorded using Leica M205 FA and Leica Application Suite X. Northern blots were quantitated using Image J
Quantitative real-time PCR
Total RNA was extracted from wing imaginal discs of late third instar larvae in TRIzol reagents (Ambion) according to the manufacturer's instructions. RNA samples were DNase treated according to manufacturer’s instructions (Thermo Fisher; AM1907) and reverse transcribed using Verso cDNA Synthesis Kit (Thermo Fisher; AB1453)..The cDNA was used to perform the qPCR reactions following the manufacturer’s instructions for the Power SYBR Green Master Mix (Thermo Fisher; 4367659). qPCR results were normalized to α-Tubulin (for Xrp1 transcripts) or CG13220 (during mRNA-Seq validation)(Ling and Salvaterra, 2011). See Table S4 for primer sequences.
Northern Analysis
Total RNA from wing imaginal discs from wandering 3rd instar larvae was prepared in TRIzol reagents (Ambion) according to the manufacturer's instructions. Northern blot analysis was based on the Odyssey Northern blot analysis protocol (LI-COR) (http://biosupport.licor.com/support). Briefly, 1-2ug of total RNA were resolved on a formaldehyde-agarose gel and transferred overnight by capillary transfer to Biodyne B Nylon membrane (Thermofisher). RNA was crosslinked onto nylon by baking at 80oC for 30 minutes and prehybridized in ULTRAhyb-Oligo buffer (Ambion). RNA biotinylated probes were used for overnight hybridization (at 55°C or 68°C). Probes were transcribed from PCR-derived templates using T7 RNA polymerase (Maxiscript T7 Transcription kit, Ambion), according to the manufacturer's instructions. Normalization controls were Actin 5C and alpha-tubulin 84B. After stringent washes and blocking with the Odyssey Blocking buffer (containing 1%SDS), the membrane was incubated with Streptavidin IRDye 800 CW (Li-CoR) and LI-COR Odyssey Infrared Imaging System was used to detect the signal.
Measurement of in vivo translation
Translation was detected by the Click-iT Plus OPP Alexa Fluor® 594 or 488 Protein Synthesis Assay Kit (Thermofisher) as described (Sanchez et al., 2016) with some modifications. Larvae were inverted in Schneider’s Drosophila medium (containing 10% heat inactivated Fetal Bovine Serum, GIbco) and transferred in fresh medium containing 1:1000 (20uM) of Click-iT OPP reagent. Samples were incubated at room temperature for 15 minutes and rinsed once with PBS. The samples were fixed in 4% formaldehyde in 1× PEM buffer (100mM Pipes, 1mM EGTA, 1mM MgCl2) for 20 min, washed once with 1× PBS and subsequently washed with 0.5% Triton in 1× PBS for 10 min and then incubated for 10 min with 3% BSA in 1× PBS. The Click reaction took place in the dark at room temperature for 30 min. Samples were washed once with the rinse buffer of the Click reaction kit, 2 minutes with 3% BSA in 1× PBS, incubated for 1 hour at room temperature with PBT (1× PBS, 0.2% Triton, 0.5% BSA) and after that incubated overnight with the primary antibodies at 4oC. Samples were washed 3× with PT buffer (1× PBS, 0.2% Triton) and the secondary antibody was added for 2 hrs in room temperature. After 3× washes with PT and 1× with 1× PBS, the samples were incubated with the Nuclear Mask reagent (1:2000) of the Click-iT kit for 30 min. After washing 2× with 1× pBS the imaginal discs were mounted in Vectashield. Confocal laser scanning images were acquired with a Leica Laser scanning microscope SP5.
mRNA-Seq
For mRNA-Seq studies, eggs laid in 4h cohorts were raised at 25°C on yeast-glucose medium supplemented with 20μg/ml tetracycline to minimize potential differences in microbiome between genotypes. Precise genotypes were, for the wild type control: w11-18(+);p{ ry+ hs-neor FRT}82B p{w+}90E/+; for RpS17/+: w11-18(y w p{ ry+ hsF}); M(3L)67C p{ w+ ubi-GFP} p{ ry+ hs-neor FRT}80B/+; for RpS3/+: w11-18(y w p{ ry+ hsF}); p{ ry+ hs-neor FRT}82B M(3R)RpS3 p{ w+ arm-LacZ}/+; for RpS3/+ Xrp1/+: w11-18(y w p{ ry+ hsF}); p{ ry+ hs-neor FRT}82B M(3R)RpS3 p{ w+ arm-LacZ}/ p{ ry+ hs-neor FRT}82B Xrp1m2-73. Third-instar larvae were harvested 24±2h before puparium formation (this corresponded to a different time after egg-laying for each genotype). From each genotype, ~50 wing discs were dissected into Trizol reagent (25 discs into 250μL) and frozen at −80°C. Following thawing and centrifugat ion (12000g 15 min 4°C), the aqueous phase was precipitated with an equal volume of isopropanol (10 min room temperature). The pellet was recovered by centrifugation (12000g 15 min 4°C), washed with RNAse-free 75% ethanol twice, and air dried (5-10 min room temperature) after removing the remaining ethanol with a filtered RNAse-free pipette tip. Samples were resuspended in RNAse-free water, analyzed by nanodrop spectroscopy and Bioanalyzer and satisfactory samples stored at −80°C prior to library construction, paired-end sequencing, mapping and alignment by Beijing Genomics Institute. Three independent replicates were analyzed for each genotype. At least 65,000,000 clean reads were obtained from every sample and at least 89% of reads were mapped from every sample.
Bioconductor DESEQ2 (Love et al., 2014) was used to identify genes expressed differentially between control and RpS17/+, between control and RpS3/+, between control and RpS3/+ Xrp1/+, and between RpS3/+ and RpS3/+ Xrp1/+. We defined genes regulated in Rp/+ genotypes as genes that were expressed differentially from control in both RpS17/+ and in RpS3/+, with probability Padj<0.1 and ∣log2fold change∣>0.5 in each case. Out of 264 such genes, 11 were discarded due to opposite regulation in the two Minute genotypes. The number of false positives that remain due to chance deviation in gene expression in the same direction in the two Minute genotypes might be expected to be of similar magnitude. Among the 253 remaining candidate Rp+/− -regulated genes, we identified Xrp1-dependent Rp+/− -regulated genes as genes expressed differentially between control and RpS3/+ but not between control and RpS3/+ Xrp1/+ or as genes expressed differentially between control and RpS3/+ and also between RpS3/+ and RpS3/+ Xrp1/+. There were 206 such genes. GO term and KEGG pathway analyses were performed using the functional annotation tool DAVID6.8(Huang da et al., 2009b, a). We considered GO terms to be significantly enriched when p<0.05 after Benjamini correction for multiple tests.
QUANTIFICATION AND STATISTICAL ANALYSES
Statistical comparisons between individual mutant and control experiments were by Student’s t-test. Details including n values, are included in Figures and Figure legends. Where data analyzed were ratios of experimental and control results eg clone sizes compared to twin-clones, values were log-transformed for statistical analysis and mean±SEM of the log transformed data reverse-transformed for presentation. Multiple sample comparison in Figure 5 was by one-way ANOVA, with the Bonferroni and Holm method to identify significant differences. Significance of differences between developmental rates (Figure 3) was evaluated using the Mann-Whitney procedure.
DATA AVAILABILITY
Primary mRNA-Seq data and DESEQ2 analyses are available from GEO (accession number GSE112864).
Supplementary Material
Drosophila Genotypes shown in Figures Relevant to Figures 1-6 and Figures S1-S5
mRNA levels (FPKM) for genes differentially expressed in both RpS3/+ and RpS17/+ wing discs compared to wild type controls Relevant to Figure 6 and Figure S5.
Highlights.
A bZip domain protein Xrp1 is expressed in cells with Rp mutations
Xrp1 controls most gene expression changes in Rp mutant heterozygotes
Xrp1 reduces translation and growth, delays development and prevents cell competition
Many effects of Rp mutations are not direct consequences of ribosome depletion
ACKNOWLEDGEMENTS
The authors thank D. Rio and S. Kurata for strains and reagents, M. Francis, L. Johnston, P. Leopold, D. Rio, J. Warner and D. Zhang for discussions, H. Buelow, S. Emmons, D. Pan and members of our laboratory for comments. Supported by a grant from the NIH (GM120451) and by the Albert Einstein College of Medicine Human Genetics Program. Drosophila strains from the Bloomington Drosophila Stock Center (supported by NIH P40OD018537) were used in this study. Confocal Imaging was performed at the Analytical Imaging Facility, Albert Einstein College of Medicine, supported by NCI cancer center support grant (P30CA013330). This paper includes data from theses partially fulfilling of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Akdemir F, Christich A, Sogame N, Chapo J, and Abrams JM (2007). p53 directs focused genomic responses in Drosophila. Oncogene 26, 5184–5193. [DOI] [PubMed] [Google Scholar]
- Armistead J, and Triggs-Raine B (2014). Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett 588, 1491–1500. [DOI] [PubMed] [Google Scholar]
- Backhaus B, Sulkowski E, and Schlote FW (1984). A semi-synthetic, general-purpose medium for Drosophila melanogaster. Drosoph Inf Serv 60, 210–212. [Google Scholar]
- Baker NE (2017). Mechanisms of cell competition emerging from Drosophila studies. Curr Opin Cell Biol 48, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, and Hafen E (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of IRS-4. Cell 97, 865–875. [DOI] [PubMed] [Google Scholar]
- Boring L, Sinervo B, and Schubiger G (1989). Experimental phenocopy of a Minute maternal-effect mutation alters blastoderm determination in embryos of Drosophila melanogaster. Developmental Biology 132, 343–354. [DOI] [PubMed] [Google Scholar]
- Boulan L, Milan M, and Leopold P (2015). The Systemic Control of Growth. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB, and Morgan TH (1923). The third-chromosome group of mutant characters of Drosophila melanogaster. Carnegie Institute Publication 327, 1–251. [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, and Rubin GM (2004). Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Pineda CM, Xin T, Boucher J, Suozzi KC, Park S, Matte-Martone C, Gonzalez DG, Rytlewski J, Beronja S, et al. (2017). Correction of aberrant growth preserves tissue homeostasis. Nature 548, 334–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Ishihara E, Miyamura N, Narumi R, Kajita M, Fujita Y, Suzuki A, Ogawa Y, and Nishina H (2016). MDCK cells expressing constitutively active Yes-associated protein (YAP) undergo apical extrusion depending on neighboring cell status. Sci Rep 6, 28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claveria C, Giovinazzo G, Sierra R, and Torres M (2013). Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500, 39–44. [DOI] [PubMed] [Google Scholar]
- Claveria C, and Torres M (2016). Cell Competition: Mechanisms and Physiological Roles. Annu Rev Cell Dev Biol 32, 411–439. [DOI] [PubMed] [Google Scholar]
- Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, and Cmejla R (2006). Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica 91, 1456–1464. [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, and Johnston L (2004). Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, and Woolford JL Jr. (2015). Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84, 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Ura H, Brandt VL, and Zwaka TP (2013). Safeguards for cell cooperation in mouse embryogenesis shown by genome-wide cheater screen. Science 341, 1511–1514. [DOI] [PubMed] [Google Scholar]
- Ellis SR (2014). Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim Biophys Acta 1842, 765–768. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, and Milkereit P (2005). Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20, 263–275. [DOI] [PubMed] [Google Scholar]
- Francis MJ, Roche S, Cho MJ, Beall E, Min B, Panganiban RP, and Rio DC (2016). Drosophila IRBP bZIP heterodimer binds P-element DNA and affects hybrid dysgenesis. Proc Natl Acad Sci U S A 113, 13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P, Shiio Y, Cheng PF, Parkhurst SM, and Eisenman RN (1996). Myc and Max homologs in Drosophila. Science 274, 1523–1527. [DOI] [PubMed] [Google Scholar]
- Golic KG (1991). Site-specific recombination between homologous chromosomes in Drosophila. Science 252, 958–961. [DOI] [PubMed] [Google Scholar]
- Hirsch CA, and Hiatt HH (1966). Turnover of liver ribosomes in fed and in fasted rats. J Biol Chem 241, 5936–5940. [PubMed] [Google Scholar]
- Hogan C, Dupre-Crochet S, Norman M, Kajita M, Zimmermann C, Pelling AE, Piddini E, Baena-Lopez LA, Vincent JP, Itoh Yv et al. (2009). Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol 11, 460–467. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Kajita M, Hogan C, Harris AR, Dupre-Crochet S, Itasaki N, Kawakami K, Charras G, Tada M, and Fujita Y (2010). Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci 123, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Ji Z, Kiparaki M, Blanco J, Rimesso G, Flibotte S, and Baker NE (2018). Ribosomal Protein S12e Has a Distinct Function in Cell Competition. Dev Cell 44, 42–55 e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Li W, Lee CH, and Baker NE (2015a). Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ 22, 1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Li W, Lee CH, and Baker NE (2015b). Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ 22, 1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, and Bassler J (2017). A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem Sci 42, 640–654. [DOI] [PubMed] [Google Scholar]
- Kucinski I, Dinan M, Kolahgar G, and Piddini E (2017). Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nat Commun 8, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson A (1998). The Minute genes in Drosophila and their molecular functions. Advances in Genetics 38, 69–134. [DOI] [PubMed] [Google Scholar]
- Lee CH, Rimesso G, Reynolds DM, Cai J, and Baker NE (2016). Whole-Genome Sequencing and iPLEX MassARRAY Genotyping Map an EMS-Induced Mutation Affecting Cell Competition in Drosophila melanogaster G3 (Bethesda: ) 6, 3207–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CT, and Brugge JS (2012). Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature 482, 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, and Baker NE (2007). Engulfment is required for cell competition. Cell 129, 1215–1225. [DOI] [PubMed] [Google Scholar]
- Lin JI, Mitchell NC, Kalcina M, Tchoubrieva E, Stewart MJ, Marygold SJ, Walker CD, Thomas G, Leevers SJ, Pearson RB, et al. (2011). Drosophila ribosomal protein mutants control tissue growth non-autonomously via effects on the prothoracic gland and ecdysone. PLoS Genet 7, e1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D, and Salvaterra PM (2011). Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One 6, e17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, and Salic A (2012). Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A 109, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, and Dawid IB (1980). Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol 138, 873–878. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FA, Herrera SC, and Morata G (2009). Cell competition, growth and size control in the Drosophila wing imaginal disc. Development 136, 3747–3756. [DOI] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, et al. (2007). The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol 8, R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, and Baserga SJ (2013). Genetics. Mysterious ribosomopathies. Science 341, 849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SN, Amoyel M, Bergantinos C, de la Cova C, Schertel C, Basler K, and Johnston LA (2014). An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 346, 1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Khincha PP, Ellis SR, Giri N, Brodie S, Chandrasekharappa SC, Donovan FX, Zhou W, Hicks BD, Boland JF, et al. (2017). Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet 54, 417–425. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, and Thomas G (1999). Drosophila S6 kinase: a regulator of cell size. Science 285, 2126–2129. [DOI] [PubMed] [Google Scholar]
- Morata G, and Ripoll P (1975). Minutes: mutants of Drosophila autonomously affecting cell division rate. Developmental Biology 42, 211–221. [DOI] [PubMed] [Google Scholar]
- Moreno E, and Basler K (2004). dMyc transforms cells into super-competitors. Cell 117, 117–129. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, and Morata G (2002). Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 416, 755–759. [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, and Johnston LA (2010). Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19, 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, and Edgar BA (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, and Dickson BJ (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851–860. [DOI] [PubMed] [Google Scholar]
- Nikolov EN, Dabeva MD, and Nikolov TK (1983). Turnover of ribosomes in regenerating rat liver. Int J Biochem 15, 1255–1260. [DOI] [PubMed] [Google Scholar]
- Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Poll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, et al. (2013). Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One 8, e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, and Glaser T (2004). Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne EM, Virgilio M, Narla A, Sun H, Levine M, Paw BH, Berliner N, Look AT, Ebert BL, and Khanna-Gupta A (2012). L-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood 120, 2214–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela M, Casas-Tinto S, Rhiner C, Lopez-Gay JM, Dominguez O, Soldini D, and Moreno E (2010). Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev Cell 19, 562–573. [DOI] [PubMed] [Google Scholar]
- Pospisilova D, Cmejlova J, Hak J, Adam T, and Cmejla R (2007). Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica 92, e66–67. [DOI] [PubMed] [Google Scholar]
- Raiser DM, Narla A, and Ebert BL (2014). The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leukemia & lymphoma 55, 491–500. [DOI] [PubMed] [Google Scholar]
- Romero-Pozuelo J, Demetriades C, Schroeder P, and Teleman AA (2017). CycD/Cdk4 and Discontinuities in Dpp Signaling Activate TORC1 in the Drosophila Wing Disc. Dev Cell 42, 376–387 e375. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, and Lehmann R (2016). Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 18, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N, Stein D, Chen K, Goltz JS, Stevens L, and DePinho RA (1997). Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proceedings of the National Academy of Sciences (USA) 94, 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P (1979). Parameters of cell competition in the compartments of the wing disc of Drosophila. Developmental Biology 69, 182–193. [DOI] [PubMed] [Google Scholar]
- Simpson P, and Morata G (1981). Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Developmental Biology 85, 299–308. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, and Hawley RS (2000). Drosophila protocols (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Tsurui-Nishimura N, Nguyen TQ, Katsuyama T, Minami T, Furuhashi H, Oshima Y, and Kurata S (2013). Ectopic antenna induction by overexpression of CG17836/Xrp1 encoding an AT-hook DNA binding motif protein in Drosophila. Bioscience, biotechnology, and biochemistry 77, 339–344. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Li W, Zhuo N, Pellock B, and Baker NE (2007). Genes affecting cell competition in Drosophila. Genetics 175, 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A, Blanc L, and Lipton JM (2014). Diamond Blackfan anemia: a model for the translational approach to understanding human disease. Expert Rev Hematol 7, 359–372. [DOI] [PubMed] [Google Scholar]
- Wagstaff L, Goschorska M, Kozyrska K, Duclos G, Kucinski I, Chessel A, Hampton-O'Neil L, Bradshaw CR, Allen GE, Rawlins EL, et al. (2016). Mechanical cell competition kills cells via induction of lethal p53 levels. Nat Commun 7, 11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, and Ready DF (1991). Cell death in normal and rough eye mutants of Drosophila. Development 113, 825–839. [DOI] [PubMed] [Google Scholar]
- Xu T, and Rubin GM (1993). Analysis of genetic mosaics in the developing and adult Drosophila tissues. Development 117, 1223–1236. [DOI] [PubMed] [Google Scholar]
- Zhang G, Xie Y, Zhou Y, Xiang C, Chen L, Zhang C, Hou X, Chen J, Zong H, and Liu G (2017). p53 pathway is involved in cell competition during mouse embryogenesis. Proc Natl Acad Sci U S A 114, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *Burke R, and Basler K (1996). Hedgehog-dependent patterning in the Drosophila eye can occur in the absence of Dpp signaling. Developmental Biology 179, 360–368. [DOI] [PubMed] [Google Scholar]
- Duffy JB, Wells J, and Gergen JP (1996). Dosage-sensitive maternal modifiers of the drosophila segmentation gene runt. Genetics 142, 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Ji Z, Kiparaki M, Blanco J, Rimesso G, Flibotte S, and Baker NE (2018). Ribosomal Protein S12e Has a Distinct Function in Cell Competition. Dev Cell 44, 42–55 e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, and Edgar BA (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, and Neufeld TP (2000). Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Drosophila Genotypes shown in Figures Relevant to Figures 1-6 and Figures S1-S5
mRNA levels (FPKM) for genes differentially expressed in both RpS3/+ and RpS17/+ wing discs compared to wild type controls Relevant to Figure 6 and Figure S5.
Data Availability Statement
Primary mRNA-Seq data and DESEQ2 analyses are available from GEO (accession number GSE112864).