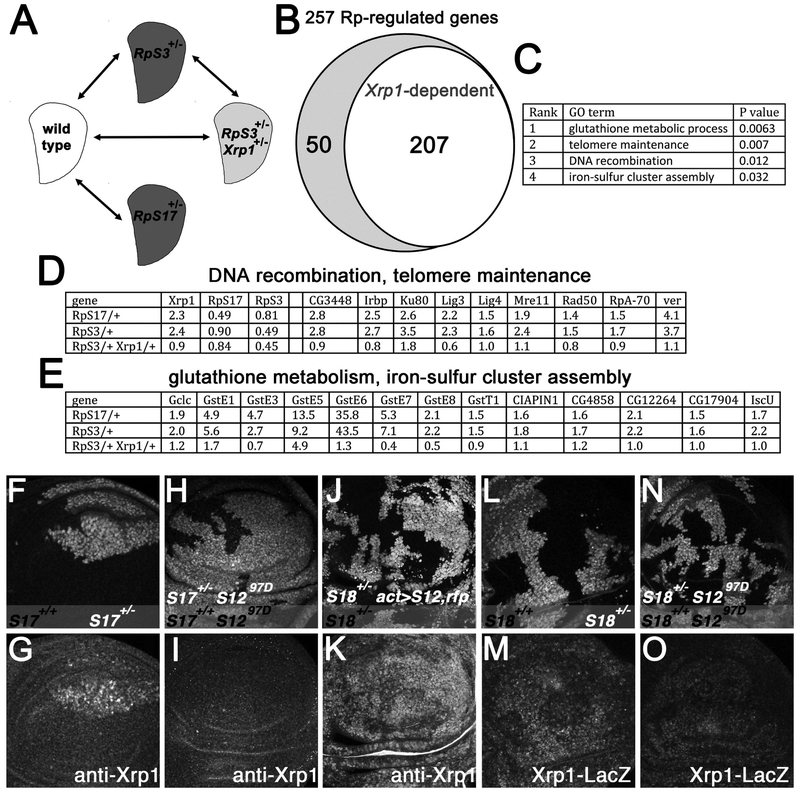
Figure 6. Xrp1 regulates many genes and is induced by RpS12.
A) mRNA was isolated from third instar wing discs enabling comparisons between four genotypes. B) Transcript levels of 253 genes were altered in both RpS17++/− and RpS3++/− and 206 of these changes were regulated by Xrp1. C) 4 GO terms for Biological Process were significantly enriched in the Rp+/− -regulated genes (ie P<0.05 after Benjamini correction). D) Expression levels (fold changes relative to wild type control according to Deseq2) for Xrp1, RpS17, RpS3 and for the 9 Rp+/−-regulated genes with telomere maintenance and DNA recombination GO terms, all of which were regulated by Xrp1. E) Expression levels (fold changes relative to wild type control according to Deseq2) for the 13 Rp+/−-regulated genes with glutathione metabolic process and iron-sulfur cluster GO terms, all of which were regulated by Xrp1. F) RpS17+/− wing imaginal disc almost entirely occupied by RpS17+/+ clones (unlabelled cells). G) Elevated Xrp1 protein levels in RpS17+/− cells. H) RpS17+/− rpS1297D/97D wing imaginal disc almost entirely occupied by RpS17+/+ rpS1297D/97D clones (unlabelled cells). I) Xrp1 protein is not elevated in the RpS17+/− rpS1297D/97D cells. J) Clonal expression of RpS12 and RFP proteins RpS18+/− wing disc. K) Xrp1 protein is elevated by RpS12 over-expression. L) RpS18+/+ clones (unlabelled cells) in RpS18+/− wing disc. M) Xrp1-LacZ is elevated in RpS18+/− cells. N) RpS18++/+ rpS1297D/97D clones (unlabelled cells) in RpS18+/− rpS1297D/97D wing disc. O) Xrp1-LacZ is not strongly elevated in the RpS18+/− rpS1297D/97D cells. Supplemental data related to this Figure is shown in Figure S5 and Table S1.