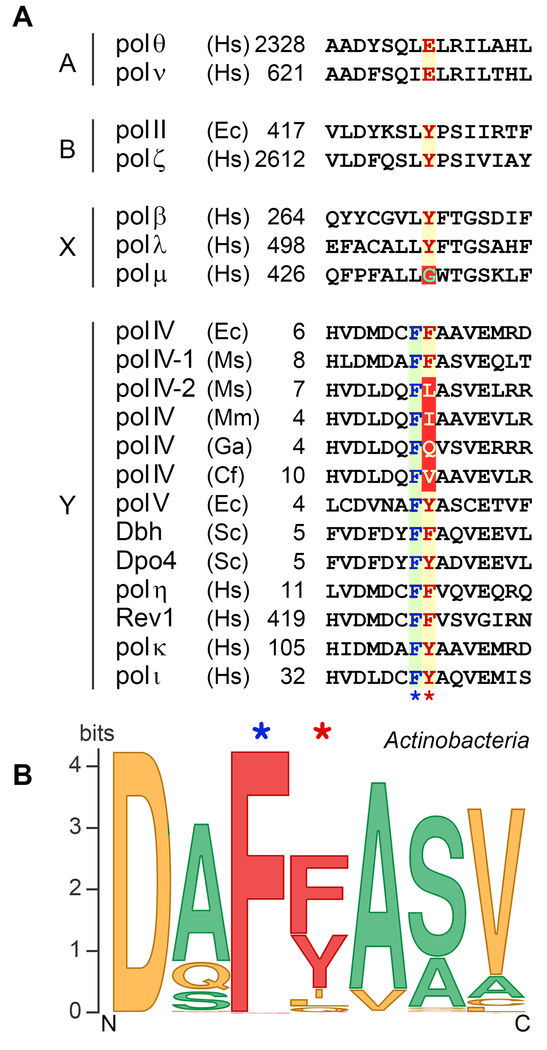
Figure 2. Multiple sequence alignments of the amino acid region involved in sugar discrimination in TLS DNA polymerases.
(A) Sequence alignments for TLS polymerases from A-, B, X, and Y families. The highly conserved single amino acid residue within each sequence responsible for the steric exclusion of ribonucleotides is shown in red letters over a yellow background and is indicated by the red star below the alignment. The unusual steric gate Gly in pol μ is indicated in green over a red background. The unconventional steric gate residues [I, L, Q and V] found in some DinB polymerases from Actinobacteria are shown in yellow letters over a red background. Almost invariant amino acid [F] juxtaposed to the steric gate residue of Y-family polymerases is shown in blue letters over a green background and is indicated by the blue star below the alignment. The numbers indicate the amino acid position of the first residue shown and are relative to the N-terminus for each polymerase. The species abbreviations are as follows: Ec, Escherichia coli; Ms, Mycobacterium smegmatis; Mm, Microbacterium mangrovi; GA, Gordonia araii; Cf, Cellulomonas fimi; Sa, Sulfolobus acidocaldarius; Ss, Sulfolobus solfataricus; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens. (B) Steric gate polymorphism among actinobacterial DinB homologs. A sequence logo for the fragment of DNA polymerase IV from Actinobacteria consisting of steric gate residue and three flanking residues on each side was created using Weblogo program (Crooks et al., 2004; Schneider and Stephens, 1990). The original sequence set consisted of 3759 actinobacteria sequences from Uniprot depository (including computationally translated sequences from TrEMBL database), predicted by InterPro signature IPR022880 as belonging to DNA polymerase IV family (retrieved on September 3, 2017, http://www.ebi.ac.uk/interpro/entry/IPR022880) (Finn et al., 2017). Sequences exceeding the threshold of 90% identity were removed, resulting in the smaller set of 1550 proteins. Multiple sequence alignment was performed using Kalign program (Lassmann et al., 2009). Sequences poorly aligned or with the gaps in the steric gate region were removed, which left 1347 proteins that were used for the sequence logo construction. Amino acids are color-coded according to their molecular weight (MW). Steric gate position in the wild type DNA polymerases are most often occupied by bulky residues (shown in red). Amino acids with lower MW identified in DinB homologs from Actinobacteria are shown in yellow. Amino acids containing the smallest side chains (shown in green) are often used to replace the steric gate residues in recombinant mutant polymerases. We thank William Taft and Iosif Vaisman (George Mason University) for creating the Dpo4 alignments and logo. A color version of the figure is available online.