Figure 1.
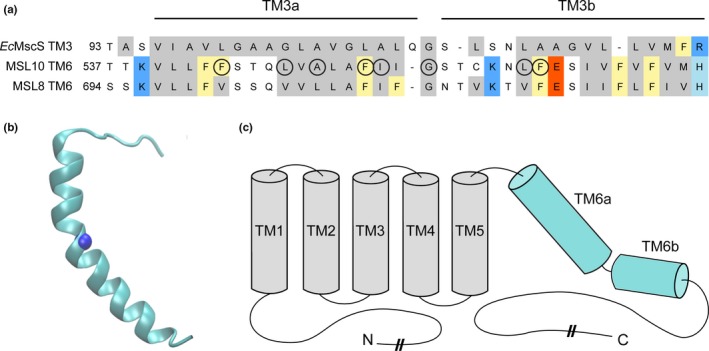
Identification of potential pore‐disrupting residues in MSL10. (a) Alignment of the pore‐lining domain of Escherichia coli MscS and corresponding regions of Arabidopsis thaliana MSL10 and MSL8. Acidic residues are indicated in red; basic residues in blue; and nonpolar in gray. Phe residues are marked yellow. Circles indicate residues that were analyzed in this report. (b) Side view of the predicted structure of the MSL10 TM6, created with I‐TASSER. The side chain at G556, predicted to form a kink, is indicated with a blue sphere. (c) Predicted topology of MSL10, indicating soluble N‐ and C‐termini and six membrane‐spanning helices. The lengths of the N‐ and C‐termini are not to scale
