Abstract
The rapid advancement of CRISPR technology has enabled targeted epigenome editing and transcriptional modulation in the native chromatin context. However, only a few studies have reported the successful editing of the epigenome in adult animals in contrast to the rapidly growing number of in vivo genome editing over the past few years. In this review, we discuss the challenges facing in vivo epigenome editing and new strategies to overcome the huddles. The biggest challenge has been the difficulty in packaging dCas9 fusion proteins required for manipulation of epigenome into the adeno-associated virus (AAV) delivery vehicle. We review the strategies to address the AAV packaging issue, including small dCas9 orthologues, truncated dCas9 mutants, a split-dCas9 system, and potent truncated effector domains. We discuss the dCas9 conjugation strategies to recruit endogenous chromatin modifiers and remodelers to specific genomic loci, and recently developed methods to recruit multiple copies of the dCas9 fusion protein, or to simultaneous express multiple gRNAs for robust epigenome editing or synergistic transcriptional modulation. The use of Cre-inducible dCas9-expressing mice or a genetic cross between dCas9- and sgRNA-expressing flies has also helped overcome the transgene delivery issue. We provide perspective on how a combination use of these strategies can facilitate in vivo epigenome editing and transcriptional modulation.
Keywords: Adeno-associated virus, cis-regulatory elements, CRISPR activation, CRISPR interference, epigenome editing, epigenetic regulation
Introduction
CRISPR (clustered regulatory interspaced short palindromic repeat)/Cas9-based RNA-guided DNA endonuclease technology has transformed research in biomedical science and engineering. It has now become the preferred platform for genome (Hsu et al. 2014; Wright et al. 2016) and epigenome editing (Pulecio et al. 2017; Thakore et al. 2016). The most widely used CRISPR system in mammalian cells consists of a single guide RNA (sgRNA) and a Cas9 nuclease (Cho et al. 2013; Cong et al. 2013; Mali et al. 2013) (Figure 1A). The sgRNA is a targeting sequence that consists of CRISPR RNA array (crRNA) and trans-activating crRNA (tracrRNA). On the other hand, Cas9 is an RNA-guided DNA endonuclease that induces a DNA double-strand break through nicking each strand of DNA by RuvC1 and HNH domains. As a general principle, sgRNA binds with Cas9 and guides the complex to cleave the target DNA. CRISPR/Cas9 system has been used to generate a gene knockout (Cho et al. 2013; Shalem et al. 2014) and knockin (Paquet et al. 2016; Sadhu et al. 2018).
Figure 1.
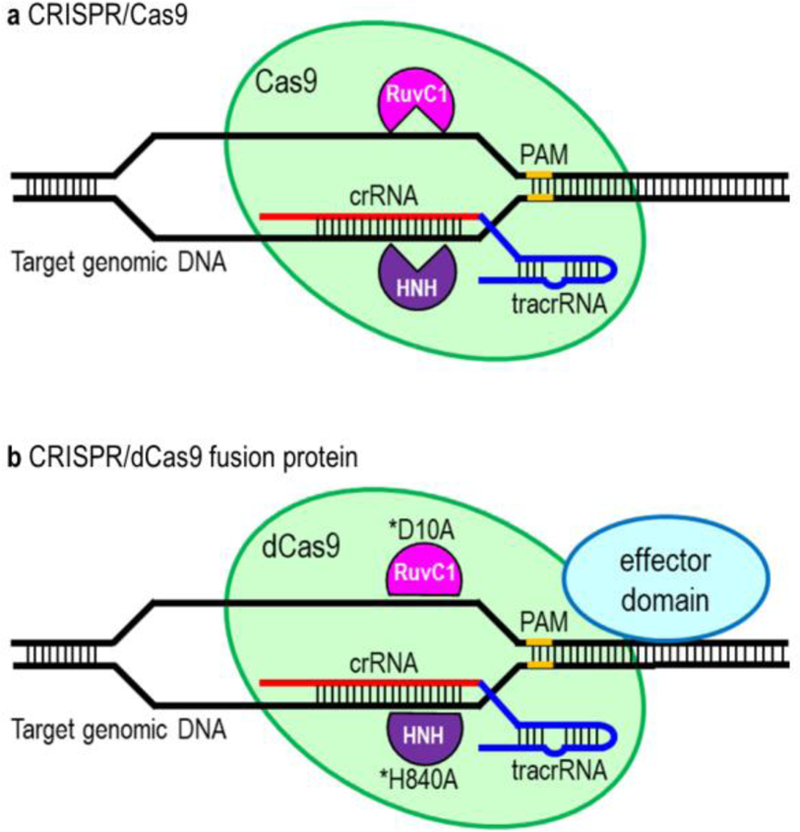
CRISPR/Cas9 and CRISPR/dCas9 systems. (A) CRISPR/Cas9 system. CRISPR/Cas9 system consists of a sgRNA (a chimeric of crRNA and tracrRNA) and Cas9. crRNA is a targeting sequence while tracrRNA functions as a Cas9 nuclease-recruiting sequence. Cas9 possess a nuclease activity to cleave the target DNA and induces a DNA double-strand break through nicking each strand of DNA by RuvC1 and HNH domains. (B) CRISPR/dCas9 fusion protein. dCas9 is nuclease-defective but posses DNA binding ability. dCas9 is derived by introducing two silencing mutations (D10A and H840A) on the RuvC1 and HNH domains, respectively. dCas9 is fused to an effector domain for targeted epigenome editing or transcriptional modulation.
More recently, the CRISPR/Cas9 has been retooled to modify epigenome and modulate transcription. The major workhorse of CRISPR-mediated epigenome editing has been the nuclease-dead Cas9 (dCas9) that allows binding of Cas9 to target DNA without inducing cleavage. dCas9 was derived by introducing two silencing mutations (D10A and H840A) on the RuvC1 and HNH nuclease domains (Qi et al. 2013) (Figure 1B). The fusion of the dCas9 to the catalytic domains of various epigenetic effectors has greatly expanded the scope of CRISPR applications to epigenome editing. These include: to modulate epigenetic marks and gene expression (Hilton et al. 2015; Kearns et al. 2015; Kwon et al. 2017; Thakore et al. 2015), manipulate nuclear architecture and chromatin loops (Hao et al. 2017; Morgan et al. 2017), and visualize chromosome organization and dynamics via chromosome imaging (Chen et al. 2013; Ma et al. 2015). In addition, stable and heritable alteration of epigenetic marks and gene expression can be achieved in post-mitotic tissues in adult animals in vivo without the need to genetically modify the DNA sequence.
Currently, adeno-associated virus (AAV) is the most commonly used viral vectors for packaging and delivery of CRISPR components in vivo (Moreno et al. 2018; Senis et al. 2014; Swiech et al. 2015; Thakore et al. 2018; Yang et al. 2016). The development of dual-vector AAV system and recent discoveries of small Cas9 orthologues (e.g. SaCas9 and CjCas9) along with truncated regulatory elements have greatly facilitated the increase in popularity of AAV-CRISPR systems for genome editing in vivo (Kim et al. 2017; Ran et al. 2015; Swiech et al. 2015). AAV is attractive for in vivo applications owing to its good safety profile, high efficacy of viral delivery and transduction, and long-term stable transgene expression in targeted tissues of a wide range of animal models (Colella et al. 2018; Mingozzi and High 2011). AAV is non-pathogenic to human, elicits a very mild immune response, and rarely cause unwanted genome integration events. However, there have been very few studies reported in vivo epigenome editing and transcriptional modulation. One of the greatest challenges in in vivo application is to package the dCas9 fusion proteins into the limited payload capacity of AAV. A fusion of dCas9 to very large effector domains is often required for robust in vivo epigenome editing. Furthermore, to achieve synergistic effects, it requires the recruitment of multiple copies of dCas9 fusion protein and co-expression of multiple sgRNAs. All these constraints have hampered the broad use of AAV-CRISPR system for epigenome editing and transcriptional modulation in adult animals.
In this review, we discuss the design principles of the CRISPR system that have been previously adapted for in vivo epigenome editing and transcriptional modulation in animals. We also summarize major CRISPR-based tools that have been developed for targeted modulation of epigenetic marks and gene expression in mammalian cells. In addition, we discuss recent advances in adeno-associated virus delivery of CRISPR fusion proteins that can facilitate in vivo application of epigenome editing tools.
CRISPR-based epigenome editing and transcriptional modulation tools
In general, the dCas9 fusion proteins allow epigenome editing and transcriptional modulation at targeted genomic loci with high specificity, minimal off-target effects and without obvious global impact (Mendenhall et al. 2013; Thakore et al. 2015). Table 1 lists CRISPR-based approaches that have been used to edit epigenetic marks and chromatin states. Despite a high target specificity observed in the majority of dCas9 fusion proteins, the potency and stability of epigenome modified by the various transcriptional modulation and epigenome editing tools vary, depending on the chromatin microenvironment and local chromatin state (Cano-Rodriguez et al. 2016). For example, dCas9-KRAB (Gilbert et al. 2013; Thakore et al. 2015) and dCas9-VP64 (Maeder et al. 2013b; Perez-Pinera et al. 2013) are commonly used transcriptional regulators to repress and activate gene expression, respectively. The effects of CRISPR activators (e.g. dCas9-VP64) and repressors (e.g. dCas9-KRAB) are usually transient, but epigenetic marks left by targeted epigenetic modifiers can be inherited by the daughter cells (Amabile et al. 2016). In addition, dCas9-VPR (Chavez et al. 2015) and dCas9-TV (Li et al. 2017) are more potent transactivation domains than the dCas9-VP64. dCas9-VPR is the fusion of dCas9 to a tripartite VP64-p65-Rta (VPR) transactivation domain. On the other hand, dCas9-TV is the fusion of dCas9 to 6TAL (six copies of the TALE transactivation domain motif) and VP128 (correspond to two VP64 moieties or 8 repeats of the VP16 motif) transactivation domains. Similar to VP64, the VPR and TV transactivation domains function to recruit endogenous transcription complexes to the dCas9 targeting sites on the promoter region. However, the gene activation efficiency of these transactivation domains is dependent on the epigenetic state of the gene. Conversely, dCas9-based epigenetic modifiers such as dCas9-P300 histone acetyltransferase directly alter chromatin states or epigenetic marks on the promoter and enhancer regions to activate gene expression (Hilton et al. 2015). dCas9-P300 exerted a stronger transcriptional activation than dCas9-VP64, despite a distinct mechanism of action between these two approaches (Hilton et al. 2015). Multiple sgRNAs and protein scaffold systems (e.g. SunTag and SAM) have also been adapted to increase the potency of epigenome editing (Huang et al. 2017; Morita et al. 2016) and transcriptional modulation (Williams et al. 2018; Xu et al. 2016) by recruiting multiple copies of dCas9 fusion proteins for synergistic effects.
Table 1.
CRISPR-based tools for epigenome editing and transcriptional modulation
| Purpose | CRISPR fusion protein | Remark | Reference |
|---|---|---|---|
| transcriptional interference |
dCas9 | repress gene expression by interfering transcriptional elongation, RNA polymerase binding, or transcription factor binding |
(Qi et al. 2013) |
| transcriptional repression |
dCas9-KRAB | induce heterochromatin formation, decrease chromatin accessibility and induce histone H3K9me3 at the enhancer and promoter regions |
(Gilbert et al. 2013; Thakore et al. 2015) |
| transcriptional repression |
dCas9-KRAB-MeCP2 | robust silencing of multiple genes | (Yeo et al. 2018) |
| transcriptional activation |
dCas9-VP64 | recruit multiple components of the transcription pre- initiation complex, including endogenous histone acetyltransferases to induce histone H3K27ac at the enhancer and promoter regions |
(Maeder et al. 2013b; Perez-Pinera et al. 2013) |
| transcriptional activation |
dCas9-VPR | VPR is a VP64-p65-Rta tripartite activator that is more potent than VP64 |
(Chavez et al. 2015) |
| transcriptional activation |
dCas9–6TAL-VP128 (dCas9-TV) |
dCas9-TV is more potent than dCas9-VP64 | (Li et al. 2017) |
| transcriptional activation |
dCas9VPH (a fusion of dCas9-VP192 and P65- HSF1 activator domains) |
induce human pluripotent reprogramming by activating endogenous master reprogramming factor genes |
(Weltner et al. 2018) |
| DNA methylation | dCas9-DNMT3A | targeted CpG methylation of the promoter silences gene expression; targeted methylation of a CTCF loop anchor site blocks CTCF binding and interferes DNA looping |
(Liu et al. 2016; Vojta et al. 2016) |
| DNA methylation | dCas9-SunTag- DNMT3A |
SunTag recruits multiple copies of antibody-fused DNMT3A to increase CpG methylation |
(Huang et al. 2017) |
| DNA methylation | dCas9-DNMT3A- DNMT3L |
multimerization of DNMT3A/DNMT3L complexes on the promoter to induce long term hypermethylation and gene silencing |
(Saunderson et al. 2017; Stepper et al. 2017) |
| DNA methylation | triple dCas9-based ETR combination |
triple engineered transcriptional repressors (ETRs) combination using dCas9-DNMT3A, dCas9-DNMT3L and dCas9-KRAB to promote long-term silencing of endogenous genes |
(Amabile et al. 2016) |
| DNA demethylation |
dCas9-TET1 | targeted demethylation of the promoter or enhancer activates gene expression or promotes active chromatin state |
(Liu et al. 2016; Liu et al. 2018b) |
| DNA demethylation |
dCas9-SunTag-TET1 | the linker length of original SunTag was changed to 22 amino acids to improve targeted demethylation efficiency by efficiently recruiting multiple copies of antibody-fused TET1 |
(Morita et al. 2016) |
| histone methylation | dCas9-PRDM9 dCas9-DOT1L |
sustain re-expression of epigenetically silenced genes by inducing H3K4me3 (using PRDM9) and H3K79me (using DOT1L) marks |
(Cano-Rodriguez et al. 2016) |
| histone methylation | dCas9-EZH2 dCas9-FOG1 |
repress chromatin state by inducing H3K27me3 marks on the promoter |
(O’Geen et al. 2017) |
| histone demethylation |
dCas9-LSD1 | remove enhancer-associated chromatin modifications (loss of H3K4me2 and H3K27ac) to downregulate target gene expression |
(Kearns et al. 2015) |
| histone acetylation | dCas9-P300 | promote robust transcriptional activation by inducing H3K27ac at the enhancer and promoter regions |
(Hilton et al. 2015) |
| histone deacetylation |
dCas9-HDAC3 | repress gene transcription by removing H3K27ac marks in the promoter |
(Kwon et al. 2017) |
| chromatin remodeling |
FIRE-dCas9 system using dCas9-MS2 anchor and Fkbp/Frb dimerizing fusion proteins |
silence (Hp1/Suv39h1 heterochromatin complex recruitment) or activate [mSWI/SNF (BAF) complex recruitment] target gene expression by recruiting endogenous chromatin regulators to specific genomic loci; rapid washout of chromatin regulators to reverse manipulation of epigenetic states |
(Braun et al. 2017) |
| chromatin remodeling |
protein trans-splicing to ligate chemical moieties to dCas9 |
dCas9-IntN and IntC-cargo (e.g. IntC-JQ1 and IntC- UNC3866) were used to recruit endogenous chromatin regulators (e.g. Brd4T bromodomain and PRC1 chromodomain) to targeted genomic loci |
(Liszczak et al. 2017) |
| chromatin looping | bivalent dCas9-Zip complexes |
induce DNA looping between the promoter and distal enhancer by heterodimerization of dSpCas9-Zip and dStCas9-Zip upon binding to two DNA target sites |
(Hao et al. 2017) |
| chromatin looping | CRISPR-dCas9 (CLOuD9) |
dSpCas9-PYL1 and dSaCas9-ABI1 induce a chromatin loop formation through dimerization of PYL1 and ABI1 domains upon addition of abscisic acid |
(Morgan et al. 2017) |
| chromatin opening | proximal CRISPR targeting |
dSpCas9 binds the proximal target sites to induce the reconfiguring and opening of the local chromatin structure |
(Chen et al. 2017) |
Note: Brd4T, bromodomain containing 4T; CLOuD9, chromatin loop reorganization using CRISPR-dCas9; DNMT3A, DNA methyltransferase 3 alpha; DOT1L, DOT1 like histone lysine methyltransferase; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; FIRE, Fkbp/Frb inducible recruitment for epigenome editing; FOG1, zinc finger protein, FOG family member 1; HDAC3, histone deacetylase 3; JQ1, a small molecule; KRAB, Krüppel associated box; LSD1, lysine demethylase 1A; MS2, bacteriophage coat proteins; P300, E1A binding protein p300; PRC1, protein regulator of cytokinesis 1; PRDM9, PR/SET domain 9; TET1, tet methylcytosine dioxygenase 1; UNC3866, a modified peptide; VPR, VP64-p65-Rta
The dCas9 has been fused to the catalytic domain of histone-modifying enzymes (e.g DNA methyltransferase and methylcytosine dioxygenase, histone methyltransferase, demethyltransferase, acetyltransferase, and histone deacetyltransferase) and chromatin-remodeling complexes (e.g. SWI/SNF complex components, bromodomain and chromodomain proteins) to modify epigenetic marks and chromatin states at the targeted cis-regulatory elements. For example, the dCas9-DNMT3A enabled transient targeted DNA methylation on the promoter (Liu et al. 2016; Vojta et al. 2016), while long-term hypermethylation and gene silencing were achieved through hit-and-run approaches using dCas9-DNMT3A-DNMT3L (Saunderson et al. 2017; Stepper et al. 2017) or a combination use of dCas9-DNMT3A, dCas9-DNMT3L and dCas9-KRAB (Amabile et al. 2016). These transient hit-and-run approaches enabled persistent de novo DNA methylation on the targeted CpG islands by instructing self-sustaining repressive epigenetic states in a chromatin environment and resisting to transcriptional activation stimuli (Amabile et al. 2016). In contrast, robust and transient DNA demethylation of the promoter to induce target gene expression was achieved using dCas9-TET1 (Liu et al. 2016; Liu et al. 2018b). TET1 is a methylcytosine dioxygenase that plays a key role in active DNA demethylation by converting 5-methylcytosines (5mCs) to 5-hydroxymethylcytosines (5hmCs) (Guo et al. 2011). Stable histone methylation was achieved through dCas9-PRDM9 and dCas9-DOT1L to re-express epigenetically silenced genes (Cano-Rodriguez et al. 2016). Both PRDM9 and DOT1L are a histone-lysine N-methyltransferase. PRDM9 methylates ‘Lys-4’ of histone H3 (Wu et al. 2013), whereas DOT1L methylates ‘Lys-79’ of histone H3 (Feng et al. 2002). Efficient histone demethylation of native enhancers has also been achieved through dCas9-LSD1 to downregulate target gene expression (Kearns et al. 2015). LSD1 (also known as KDM1A) is a lysine-specific histone demethylase that acts as a transcriptional corepressor by mediating demethylation of H3K4me (Shi et al. 2004). In addition, efficient, transient histone acetylation and deacetylation at the targeted enhancer and promoter regions were achieved by using dCas9-P300 (Hilton et al. 2015) and dCas9-HDAC3 (Kwon et al. 2017), respectively. P300 is a histone acetyltransferase that acts as a co-activator by mediating acetylation of H3K27ac on the enhancer region (Raisner et al. 2018). In contrast, HDAC3 functions as a corepressor by deacetylating the H3K27ac on enhancer elements (Hatzi et al. 2013). Recently, CRISPR technology has also been successfully used to recruit endogenous chromatin regulators to targeted genomic loci (Braun et al. 2017; Liszczak et al. 2017) or manipulation of nuclear architecture and chromatin looping (Hao et al. 2017; Morgan et al. 2017). However, most of these studies have not yet been tested in vivo.
In vivo CRISPR-based epigenome editing and transcriptional modulation in animals
In general, AAV was the only viral vector used for in vivo delivery of CRISPR components in adult mice. Table 2 lists all studies that reported the use of CRISPR technology for in vivo epigenome editing and transcriptional modulation. Other viral vectors such as lentivirus (Joung et al. 2017; Klann et al. 2017; Stover et al. 2017; Thakore et al. 2015) and adenovirus (Voets et al. 2017) have a larger packaging capacity than AAV to accommodate CRISPR fusion proteins, but the non-AAV vectors were used only for transduction into the cell lines or primary cells. Lentivirus is particularly useful for genome-wide screens and transduction of difficult-to-transfect primary cell (Joung et al. 2017; Koike-Yusa et al. 2013). Adenovirus is commonly used for vaccine production and cancer immune therapy (Vellinga et al. 2014). Compared with AAV, these viruses are inefficient for in vivo applications owing to high immunogenicity. AAV8 (Thakore et al. 2018; Zhou et al. 2018) and AAV9 (Chew et al. 2016; Liao et al. 2017) were used to package and deliver CRISPR components in all the previous studies. For example, a fusion of dSaCas9 and KRAB repressor domain has a transgene size that falls within the AAV packaging capacity. Using a dual-vector AAV8 system, a recent study has packaged the dSaCas9-KRAB fusion proteins and a sgRNA into two separate AAV vectors for in vivo gene silencing (Thakore et al. 2018). It is also possible to fuse dSaCas9 to the VP64 transactivation domain for packaging into the AAV vector for in vivo target gene activation. Nonetheless, the AAV vector cannot accommodate a larger and more potent dSaCas9 fusion proteins, such as a fusion of dSaCas9 to the tripartite VP64-p65-Rta (VPR) transactivation domain. Owing to the large size of VPR, fusion proteins of the VPR to the small catalytically inactive Cas9 ortholog such as CjCas9 (Kim et al. 2017) or SaCas9 (Ran et al. 2015) still are not capable of circumventing the transgene packaging issue.
Table 2.
In vivo epigenome editing and transcriptional modulation using CRISPR technology
| Purpose | CRISPR strategy | Delivery method | Significance | Animal | Reference |
|---|---|---|---|---|---|
| DNA methylation |
targeted CpG methylation using dCas9-MQ1 (a fusion between dCas9 and prokaryotic DNA methyltransferase MQ1) |
induce targeted CpG methylation in mice by zygote microinjection of dCas9-MQ1 |
specific, rapid and efficient strategy to achieve locus- specific cytosine modifications in the genome in vivo |
mouse embryo |
(Lei et al. 2017) |
| DNA demethylation |
dCas9-peptide repeat was used to recruit multiple copies of scFv-TET1 catalytic domain fusion for amplification of DNA demethylation |
in utero electroporation of all-in-one vectors |
in vivo targeted demethylation in the brain of mouse fetuses |
mouse fetus |
(Morita et al. 2016) |
| gene activation | induce trans-epigenetic remodeling by co-introducing a truncated 14bp guide RNA (dgRNA) and an MS2- P65-HSF1 (MPH) transcriptional activation complex into Cas9- or dCas9-expressing mice |
AAV9 delivery of dgRNA and MPH via intramuscular, facial vein, intra-cerebral, and tail-vein injections |
ameliorate disease phenotypes for type I diabetes, acute kidney injury, and muscular dystrophy |
adult mouse |
(Liao et al. 2017) |
| gene activation | a Cre-dependent SunTag-p65-HSF1 (SPH) transgenic mouse that stably expressing dCas9-10xGCN4 of SunTag and p65-HSF1 of SAM |
AAV8 delivery of Cre and sgRNAs via stereotactic injection |
directly converted astrocytes into functional neurons in the brain by activating endogenous neurogenic genes |
adult mouse |
(Zhou et al. 2018) |
| gene activation | a split Cas9 AAV system, which Cas9 N-terminal lobe fused to the N- split intein (Cas9N) while the C- terminal lobe fused to the C-split intein (Cas9C); Cas9C was then fused to the tripartite VPR transactivation domain |
AAV9 delivery of Cas9C- VPR and Cas9N-gRNAs via intramuscular injection |
enable dual AAV delivery of Cas9-VPR fusion proteins for gene activation in the muscle |
adult mouse |
(Chew et al. 2016) |
| gene activation | dCas9-VPR system coupled with Gal4-UAS activation to induce dominant phenotypes in vivo |
flies of the genotype dCas9-VPR were crossed to homozygous sgRNA flies |
first demonstration of dCas9-based activation in a multicellular animal |
Drosop hila |
(Lin et al. 2015) |
| gene activation | dCas9-VPR system coupled with a genome-wide collection of flies expressing sgRNAs |
flies of the genotype dCas9-VPR were crossed to a collection of flies expressing sgRNAs |
generated strong gain-of- function phenotypes in multiple tissues in vivo for large-scale genetic screens |
Drosop hila |
(Ewen- Campen et al. 2017) |
| gene activation | a flySAM1.0 system consisted of dCas9-VP64, MCP-p65-HSF1 and sgRNA-luc was used for in vivo CRISPRa luciferase assay; a flySAM2.0 all-in-one system was used for tissue-specific CRISPRa with a single genetic cross |
flySAM2.0 lines were crossed to a Gal4 line |
improve potency, scalability, and ease of use for systematic overexpression genetic analysis and screens |
Drosop hila |
(Jia et al. 2018) |
| gene activation | a Cre-inducible CRISPRa system (dCas9-SunTag) for hepatocyte- specific gene activation in the liver |
AAV delivery of Cre in dCas9-expressing mice, followed by hydrodynamic injection of plasmid pools (Fah, TA and gRNA) |
enable parallel and combinatorial genetic screening for drivers and suppressors of tumor initiation and proliferation in live animals |
adult mouse |
(Wangenst een et al. 2017) |
| gene activation | an optogenetic far-red light (FRL)- activated CRISPR-dCas9 system (FACE) based on dCas9, hybrid transactivator MS2-p65-HSF1, the bacterial phytochrome BphS and FRL illumination; in the presence of FRL, BphS is activated to convert GTP into c-di-GMP. Increased cytosolic c- di-GMP production dimerizes p65- VP64-NLS-BldD to induce expression of MS2-p65-HSF1, which are further recruited by the MS2 box of the sgRNA-dCas9 complex to activate target gene expression. |
plasmid electroporation of FACE system into mouse muscles, followed by illumination with FRL |
promote differentiation of induced pluripotent stem cells into functional neurons via precise spatiotemporal control of endogenous gene expression and trans- epigenetic remodeling |
adult mouse |
(Shao et al. 2018) |
| gene activation and repression |
dCas9-VP64 mediated transcriptional perturbation |
tail-vein injection of B- ALL cells expressing dCas9-VP64 and a custom sgRNA library |
interrogate tumor phenotypes in vivo for modeling cancer progression and therapeutic relapse |
adult mouse |
(Braun et al. 2016) |
| gene activation and repression |
a modular dual-AAV split-dCas9 system consisting of dCas9-based transcriptional regulation modules (VP64, RTA and p65 activators) (KRAB, DNMT3A, DNMT3L or FOG1 repressor) |
AAV delivery of AAV- split-KRAB-dCas9-Nrl into mouse retina |
enable in situ gene therapy by targeting disease in a genomically scarless and reversible manner |
adult mouse |
(Moreno et al. 2018) |
| gene silencing | a dual-vector AAV8 system, which two AAV vectors separately express a dSaCas9-KRAB repressor and a sgRNA |
AAV8 delivery of a dSaCas9-KRAB and a sgRNA via tail-vein injection |
reduce serum Pcsk9 and cholesterol levels in the liver |
adult mouse |
(Thakore et al. 2018) |
| gene silencing | dCas9-KRAB with a pseudotarget fishing strategy to achieve superior targeting specificity without detectable off-target activity |
lentiviral delivery of dCas9-KRAB-U6-sgRNA via stereotactic injection |
enable rapid and accurate gene function investigation in the mammalian brain |
adult mouse |
(Zheng et al. 2018) |
| histone demethylation, gene activation and repression |
dCas9-LSD1 demethylase, dCas9- KRAB repressor, or dCas9-VP64 that incorporates the synergistic activation mediator (SAM) system |
electroporation of a dCas9- effector and multiple sgRNAs |
probe gene regulatory interactions during early neural crest development |
chicken embryo |
(Williams et al. 2018) |
Note: Fah, fumarylacetoacetate hydrolase; HSF1, heat shock factor 1; KRAB, Krüppel associated box; LSD1, lysine-specific histone demethylase 1A; MS2, viral RNA stem-loop motifs; Pcsk9, proprotein convertase subtilisin/kexin type 9; SAM, synergistic activation mediator; scFv, single-chain variable fragment; TA, transcriptional activator; TET1, methylcytosine dioxygenase 1; UAS, upstream activating sequence; VPR, VP64-p65-Rta.
One of the strategies to enable AAV delivery of Cas9-VPR fusion proteins and truncated sgRNAs was to use a split-Cas9 system (Chew et al. 2016). In this system, Cas9 was split at its disordered linker (V713-D718) to generate 2.5kb N-terminal lobe with N-split-intein (Cas9N) and 2.2kb C-terminal lobe with C-split-intein (Cas9C) (Figure 2). This split-intein protein trans-splicing strategy allows seamless reconstitution of full-length Cas9, preserves Cas9 structure and function upon co-expression of these two lobes in vivo. The Cas9C was then fused to 1.6kb tripartite VPR transactivation domain (~3.8kb Cas9C-VPR) for packaging into the AAV vector. Meanwhile, the Cas9N was linked to the truncated gRNAs for packaging into another AAV vector. The use of truncated gRNAs enables nuclease-active Cas9 to bind to the genomic loci without inducing DNA double-strand breaks. This AAV-split-Cas9 system has been successfully used to activate endogenous genes in postnatal mice (Chew et al. 2016). In this study, AAV9-Cas9C-VPR and AAV9-Cas9N-gRNA were individually packaged and co-delivered to the mice by the AAV9 vectors. A similar split-Cas9 system can be designed to enable AAV delivery of dCas9 fusion proteins and full-length 20nt sgRNAs. For example, in vivo gene repression in mice has been achieved using the AAV-split-KRAB-dCas9 system (Moreno et al. 2018). However, the use of two separate AAV vectors to co-express split Cas9 or dCas9 fragments may reduce the delivery and gene modulation efficiency.
Figure 2.
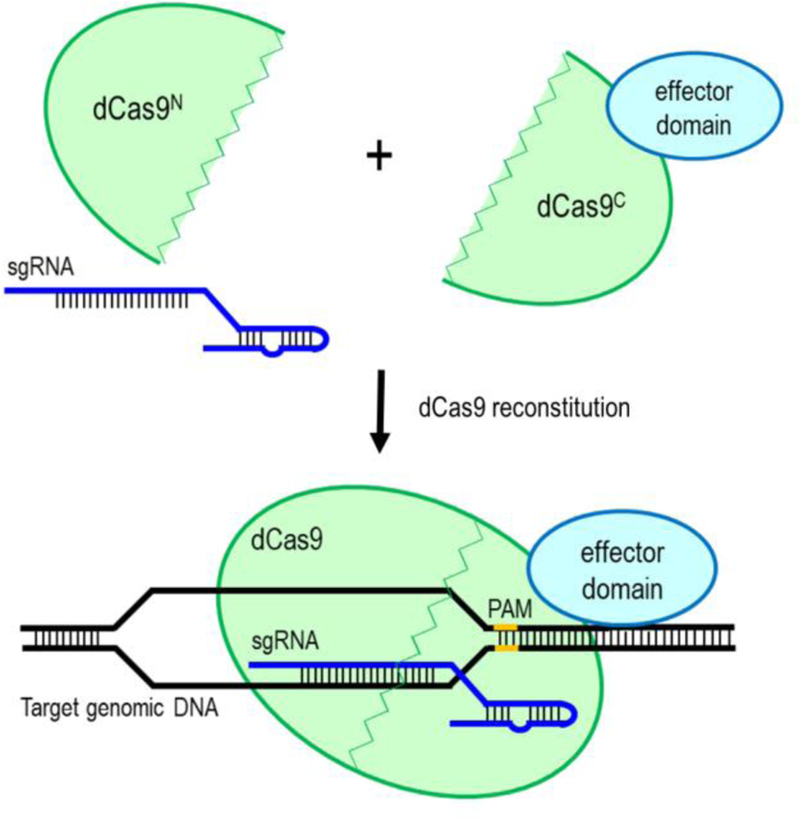
Split-dCas9 system using dual-AAV vectors. dCas9 is split into two parts, N-terminal lobe with N-split-intein (Cas9N) and C-terminal lobe with C-split-intein (Cas9C). Cas9C is then fused to an effector domain, while Cas9N is linked to the truncated gRNAs for packaging into two separate AAV vectors. Co-delivery and co-expression of these two AAV vectors in vivo will reconstitute the full-length and function of dCas9 fusion protein.
To overcome the delivery challenges associated with the large sizes of dCas9-fused activators and to enable robust in vivo gene activation, two independent studies have developed novel strategies that enable AAV delivery of Cre recombinase and sgRNAs into Cre-inducible dCas9 activator-expressing mice (Wangensteen et al. 2017; Zhou et al. 2018) (Figure 3A). One of the studies has generated a Cre-dependent SunTag-p65-HSF1 (SPH) transgenic mice by replacing VP64 in SunTag (dCas9–10xGCN4) with p65-HSF1 of synergistic activation mediator (SAM) (Zhou et al. 2018). AAV8 was then used to deliver the Cre and sgRNAs into the SPH transgenic mice. This highly potent dCas9 activator system allowed in vivo direct conversion of astrocytes into functional neurons by inducing the endogenous expression of neurogenic transcription factors. It also enabled simultaneous activation of multiple genomic loci in vivo for modulating complex genetic networks in the intact brain. In another study, a novel in vivo CRISPR activation platform was developed for parallel and combinatorial genetic screens in adult mice (Wangensteen et al. 2017). In this system, Cre-inducible dCas9 activator-expressing mice were generated by integrating dCas9-SunTag transgene into the Rosa26 locus, followed by delivery of Cre recombinase using AAV for activation of dCas9-SunTag expression in a specific tissue. Finally, pools of Sleeping Beauty transposon plasmids expressing a transcriptional activator and a library of unique sgRNAs were introduced into the mice via hydrodynamic injection (Wangensteen et al. 2017). This in vivo CRISPR activation system has enabled high-throughput screening of driver and suppressor genes involved in tumorigenesis and metastasis.
Figure 3.
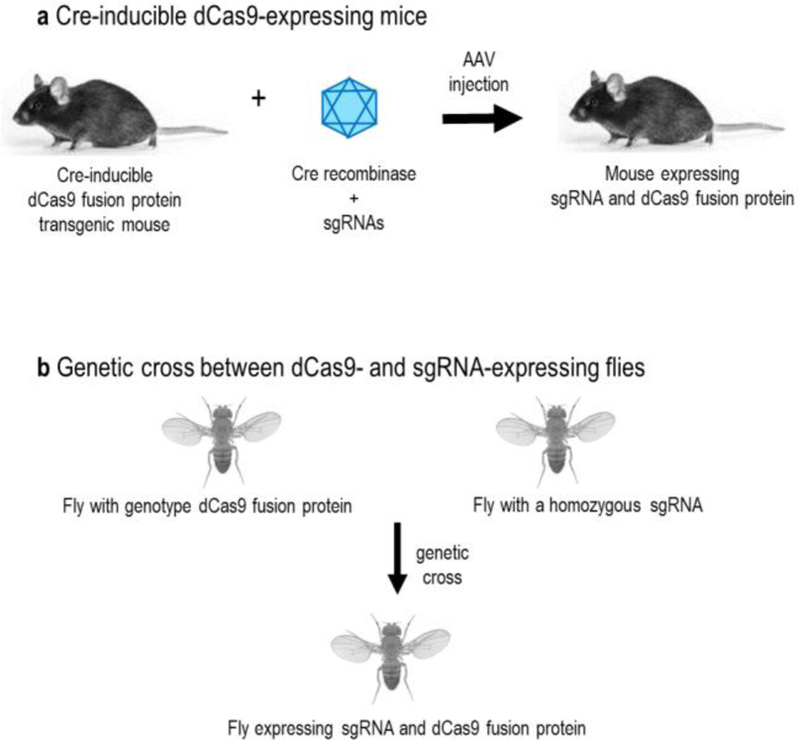
Epigenome editing and transcriptional modulation using transgenic mice or flies. (A) Cre-inducible dCas9-expressing mice. To express the sgRNA and dCas9 fusion protein in vivo, Cre recombinase and sgRNAs are packaged into the adeno-associated virus (AAV) vectors for co-delivery into the Cre-inducible dCas9 fusion protein transgenic mouse. (B) Genetic cross between dCas9- and sgRNA-expressing flies. To express sgRNA and dCas9 fusion protein in vivo, the dCas9 fusion protein and a homozygous sgRNA transgenic flies are genetic crossed.
In another elegant study, trans-epigenetic remodeling approach was developed to achieve a potent in vivo target gene activation and phenotypic change (Liao et al. 2017). In this strategy, a truncated guide RNA (dgRNA) and an MS2-P65-HSF1 (MPH) transcriptional activation complex were packaged into two separate AAV vectors for co-delivery into Cas9- or dCas9-expressing mice. Because dCas9 was not fused together with the transcriptional activator domains, the dgRNA was equipped with two MS2 domains to recruit MPH transcriptional activation complex. This approach has been successfully used to ameliorate disease phenotypes for type I diabetes, acute kidney injury, and muscular dystrophy by remodeling epigenetic marks and activating endogenous genes in transgenic mice. While AAV remains the preferred choice for delivery of CRISPR components in adult animals, electroporation has enabled introduction of an all-in-one plasmid construct consisting of a very large dCas9 fusion proteins and multiple sgRNAs for robust target gene activation, gene repression, and histone and DNA demethylation in mouse fetus (Morita et al. 2016) and chicken embryo (Williams et al. 2018).
Using Drosophila models, three independent studies have developed robust in vivo CRISPR activation systems via genetic crosses between dCas9 activator- and sgRNA-expressing flies (Ewen-Campen et al. 2017; Jia et al. 2018; Lin et al. 2015) (Figure 3B). The first demonstration of dCas9-based activation in a multicellular animal was achieved through the genetic crossing of flies with genotype dCas9-VPR and a homozygous sgRNA (Lin et al. 2015). This approach was successfully adapted to induce dominant phenotypes in vivo via the Gal4-UAS activation system. The Gal4 transcription activator was used to drive expression of the dCas9-VPR by binding to UAS enhancer regions. To enable high throughput and systematic overexpression genetic analysis, a genome-wide collection of flies expressing sgRNAs was subsequently generated for crossing with the dCas9-VPR transgenic flies (Ewen-Campen et al. 2017). This approach enabled the generation of easily recognizable gain-of-function phenotypes in multiple tissues in vivo for facilitating large-scale genetic screens.
More recently, a fly SAM system was developed to improve the effectiveness, scalability, multiplexity, and simplicity of existing CRISPR activation strategies (Jia et al. 2018). Two versions of flySAM, namely, flySAM1.0 and flySAM2.0 were developed. In the flySAM1.0 system, dCas9-VP64 and MCP-p65-HSF1 were separated by a T2A self-cleaving peptide. This was to ensure lower expression of MCP:p65-HSF1 for minimizing lethality and improving survival of flies. This flySAM1.0 line was then crossed to the sgRNA-expressing line for activating target genes in vivo. The flySAM1.0 allowed generation of stronger phenotypes than the previously established dCas9-VPR system. In fact, flySAM1.0 was able to recapitulate the previously reported overexpression phenotypes by using only a single sgRNA. By co-expressing multiple sgRNAs, flySAM1.0 enabled simultaneous activation of multiple genes in vivo. To simplify the experimental use of flySAM, flySAM2.0 was then developed through the creation of a single vector encoding both the UAS:dCas9-activator and the sgRNA. A single genetic cross between flySAM2.0 and Gal4 lines was sufficient to activate target genes in a specific tissue. The flySAM2.0 with a single sgRNA was also more potent than the dCas9-VPR with two sgRNAs. In the future, similar genetic crosses can be adapted in mice for inducible overexpression of dCas9 fusion proteins and large-scale genetic screens without the need to use AAV vectors.
Recent advances in AAV delivery of CRISPR fusion protein
Although many of the newly developed AAV-CRISPR systems have yet to be tested in vivo, these systems have enabled AAV delivery of CRISPR components for epigenome editing and transcriptional modulation in cell lines. Recent discoveries of the small Cas orthologues (Cas13d, CjCas9, and SaCas9), development of split-dCas9 (dCas9N and dCas9C), truncated Cas mutants (mini-SaCas9 and RCas9) and effector domains (mini-VPR), and RNA targeting strategies have greatly facilitated the applications of AAV-CRISPR system for epigenome editing and transcriptional modulation (Table 3).
Table 3.
Recent advances in AAV delivery of CRISPR fusion protein
| Purpose | AAV packaging strategy | Remark | Reference |
|---|---|---|---|
| gene activation |
a mini-SaCas9-VTR was created by fusing a truncated SaCas9 (mini-SaCas9) to a downsized tripartite VPR transactivation domain (VTR) |
mini-SaCas9 is nuclease-deficient but retains DNA binding activity |
(Ma et al. 2018) |
| gene activation |
an AAV-compatible CRISPR activator was designed by fusing dSaCas9 to the truncated VPR activator domain |
the truncated VPR domain has a comparable activation activity to the full-length VPR activator |
(Vora et al. 2018) |
| gene activation |
Cas9 was split into 2.5kb N-terminal lobe with N-split- intein (Cas9N) and 2.2kb C-terminal lobe with C-split-intein (Cas9C); Cas9C was then fused to the tripartite VPR transactivation domain (Cas9C-VPR); Cas9N and truncated gRNAs were constructed as a single vector (Cas9N-gRNAs) |
split-Cas9 is fully active and efficient as the full-length Cas9 |
(Chew et al. 2016) |
| gene activation |
a trans-epigenetic modulation system that uses two AAV vectors to separately package AAV-SpCas9 and AAV- dgRNA-MPH; dgRNA is a truncated 14bp guide RNA with MS2 loops, while MPH is a MS2-P65-HSF1 transcriptional activation complex |
gene activation activity of AAV-SpCas9 is similar to AAV-dSpCas9, but the use of AAV-dSpCas9 can minimize the concern of generating DNA double- strand breaks |
(Liao et al. 2017) |
| gene activation |
co-delivery of dCas9-VP64, and multiple gRNAs linked with self-cleaving ribozymes and/or tRNA for synergistic transcription activation; multiple gRNAs were simultaneously expressed from a single U6 promoter |
adenovirus was used for transduction in vivo; theoretically, it is possible to use a dual-vector AAV system to separately package dCas9-VP64 and ribozymes- linked gRNAs |
(Xu et al. 2016) |
| gene silencing |
a dual-vector AAV8 system that packages a dSaCas9- KRAB repressor and a sgRNA separately into two AAV vectors |
enable a durable, long-term target gene silencing in post-mitotic tissues in live animal |
(Thakore et al. 2018) |
| gene knockdown |
CjCas9 can be easily packaged into the AAV for targeting and cleaving complementary endogenous mRNAs in a crRNA-dependent manner |
CjCas9 is the smallest Cas9 characterized to date; RNA cleavage required the HNH domain of CjCas9, but was independent of a PAM |
(Dugar et al. 2018) |
| gene knockdown |
CjCas9 and SaCas9 can be easily packaged into the AAV for mediating PAM-independent RNA-guided RNA cleavage |
CjCas9 (2.95kb) and SaCas9 (3.16kb) are smaller than the SpCas9 (4.10kb); AAV delivery is yet to be demonstrated |
(Strutt et al. 2018) |
| gene knockdown |
Cas13d orthologs, including EsCas13d and RspCas13d can be easily packaged into the AAV for targeting and cleaving endogenous mRNA |
Cas13d is 190–300 amino acids smaller than that of Cas13a-Cas13c; AAV delivery is yet to be demonstrated |
(Yan et al. 2018) |
| gene knockdown |
a RCas9 system was developed by fusing a truncated dCas9 to the PIN RNA endonuclease domain from SMG6; 4.3kb PIN-dCas9(ΔHNH) or 3.9kb PIN-dCas9(ΔHNH, ΔREC2) was used for eliminating toxic microsatellite repeat expansion RNA |
the PIN-dCas9(ΔHNH) lacks the HNH domain but has a normal protein folding and an equivalent enzymatic activity to the full-length Cas9 |
(Batra et al. 2017) |
| gene knockdown and alternative splicing |
a RNA-targeting Cas13d (CasRx) was packaged into the AAV for efficient knockdown of mRNA transcripts; AAV delivery of catalytically inactive dCasRx splice effectors and a three-guide array to target cis elements of pre-mRNA for manipulating alternative splicing |
CasRx has a size smaller than any known Cas9 (including CjCas9 and SaCas9) and other Cas13 classes (Cas13a-c) |
(Konermann et al. 2018) |
| gene knockdown and RNA editing |
a small size dCas13b-ADARDD was generated by fusing a truncated dCas13 to ADAR2DD(E488Q) for directing adenosine to inosine deaminase activity |
the C-terminal truncated dCas13 was still functional and small enough to package together with the ADAR2DD into a single AAV vector |
(Cox et al. 2017) |
Note: ADAR, adenosine deaminases acting on RNA; KRAB, Krüppel associated box; PAM, protospacer adjacent motif; VPR, VP64-p65-Rta.
Cas13d is the smallest class 2 CRISPR effector characterized to date (Konermann et al. 2018; Yan et al. 2018). Cas13d (~930 amino acids) is 190–300 amino acids smaller than that of Cas13a-Cas13c. It is also smaller than the compact Cas9 orthologues such as CjCas9 (984 amino acids, 2.95kb) (Kim et al. 2017) and SaCas9 (1053 amino acids, 3.16kb) (Ran et al. 2015). This remarkably small size of Cas13d effectors enabled the design of minimal Cas13d fusion proteins that can be accommodated into the AAV vectors (Figure 4A). Cas13d possesses dual RNase activities due to the presence of two HEPN motifs. Cas13d can be reprogrammed to target and cleave RNAs with minimal targeting constraints (Yan et al. 2018). One of the human codon-optimized Cas13d orthologs, CasRx has been successfully used to knockdown mRNA transcripts in human cells, with a higher efficiency and specificity than the RNA interference (shRNA) (Konermann et al. 2018). A catalytically dead Cas13d (dCas13d) that can target specific RNA elements has also been created by introducing R295A, H300A, R849A, and H854A mutations to inactivate the positively charged catalytic residues of the HEPN motifs. The dCas13d was then fused to the catalytic domains of effectors for transcriptional modulation. For example, dCasRx splice effector was generated by fusing dCasRx to the Gly-rich C-terminal domain of heterogeneous nuclear ribonucleoprotein a1 (hnRNPa1). The dCasRx splice effector was then packaged into the AAV vectors for tuning alternative splicing and correction of protein isoform ratios in postmitotic neurons of frontotemporal dementia (Konermann et al. 2018). Although has not yet been demonstrated, it is also possible to package CjCas9 and SaCas9 into the AAV vectors for targeting and cleaving complementary endogenous mRNAs independent of a PAM (Dugar et al. 2018; Strutt et al. 2018). In this case, CjCas9 or SaCas9 nuclease alone is sufficient to alter the mRNA levels by directly targeting the RNA transcripts. This way the researcher can avoid using a large dCjCas9 or dSaCas9 fusion protein to modulate the cis-regulatory elements in the genome.
Figure 4.
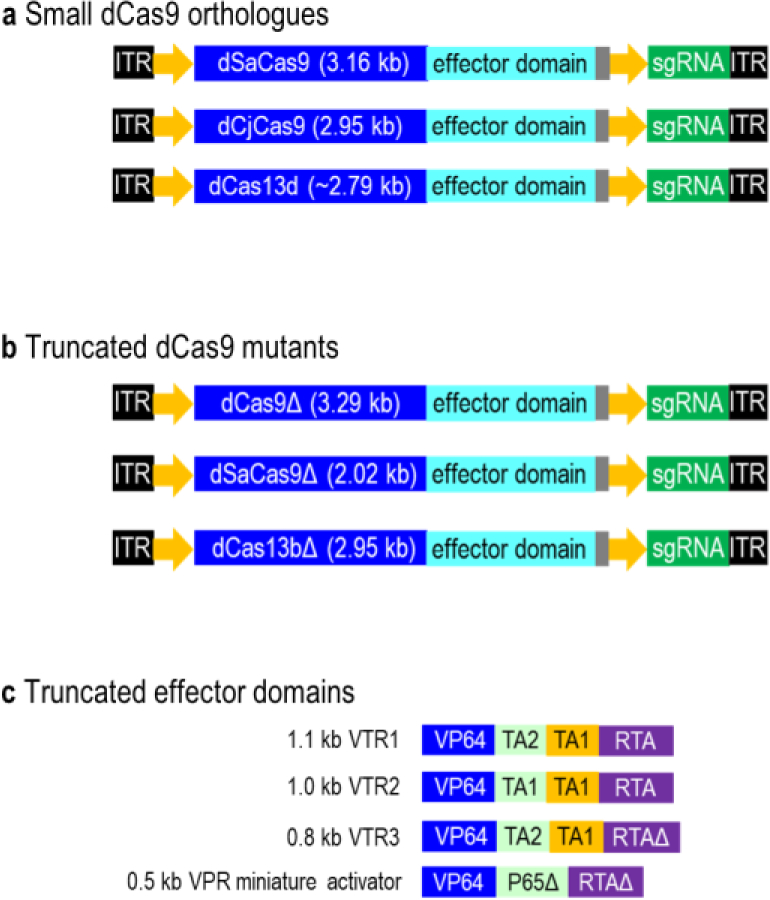
AAV-compatible dCas9 fusion proteins are derived from small dCas9 orthologues, truncated dCas9 mutants or effector domains. (A) Small dCas9 orthologues. Small Cas orthologues (dSaCas9, dCjCas9 and dCas13d) are AAV-compatible and smaller than the dSpCas9 (4.10 kb). (B) Truncated dCas9 mutants. By deleting HNH and REC2 domains of dCas9, the truncated dCas9 (reduce 0.81kb) lacks the nuclease activity but retains normal protein folding and function. By deleting conserved functional domains of dSaCas9, the truncated dSaCas9 (reduce 1.14kb) is nuclease-defective but retains DNA binding activity. Truncated dCas13b (reduce 0.32kb) lacks a C-terminal domain but retains programmable RNA binding capability. (C) Truncated effector domains. A set of compact transactivation domains is generated by removing the DNA binding domains in P65 and RTA of VPR. 1.1kb VTR1 (reduce 0.4kb) is generated by substituting the P65 domain in the VPR with the TA1 and TA2 transactivation domains. 1.0kb VTR2 (reduce 0.5kb) is created by replacing the P65 domain in the VPR with two repeats of the TA1. The partial RTA domain in the 1.1kb VTR1 is removed to generate a 0.8kb VTR3 (reduce 0.7kb). 0.5kb VPR miniature activator (reduce 1.0kb) is generated by truncating the P65 and RTA domains.
To further reduce the size of these small Cas proteins for fusing to a larger effector domain, Cas proteins were truncated without disrupting the protein folding and function (Figure 4B). For instance, dCas13b-ADARDD was created by fusing a truncated dCas13b to ADAR2DD(E488Q) for RNA editing (Cox et al. 2017). The C-terminal truncated dCas13b retains programmable RNA binding capability, while adenosine deaminase acting on RNA type 2 (ADAR2) directs adenosine to inosine deaminase activity on mRNA transcripts. Another study has developed an AAV-compatible RNA-targeting Cas9 (RCas9) system for efficient elimination of microsatellite repeat expansion RNAs by fusing a truncated dCas9 to the PIN RNA endonuclease domain from SMG6 (Batra et al. 2017). The truncated dCas9 lacks an HNH domain or the HNH and REC2 domains but retains normal protein folding and function. Alternatively, intein-mediated split dSaCas9 reconstitution system enabled the introduction of a large dSaCas9 fusion proteins into the cells through two separate AAV vectors. Using the similar concept as described for the SpCas9-based split system (Chew et al. 2016), the dSaCas9 was split at residue 739 into dSaCas9C and dSaCas9N (Ma et al. 2018). The dSaCas9C-VPR and dSaCas9N-gRNA were then packaged into two separate AAV vectors for co-expression.
Instead of truncating the dSaCas9, a recent study has truncated the VPR activation effector to reduce the size of the dSaCas9-VPR fusion protein from 5.0kb to 4.3kb (Vora et al. 2018) (Figure 4C). VPR consists of a fusion of VP64, p65 and Rta domains. Two smallest truncated VPR domains were identified through serial truncations of p65 and Rta from either the N or C-terminus. These truncated VPR domains have a comparable activation activity to the full-length VPR activator. The dSaCas9-VPR miniature activator was also significantly more robust than the previously established dCas9-VP64 of the SAM system (Konermann et al. 2015). Another example of effective AAV packaging strategy was the adaption of a mini-SaCas9-VTR by fusing a truncated SaCas9 (mini-SaCas9) to a downsized tripartite VPR transactivation domain (VTR) (Ma et al. 2018). By deleting conserved functional domains of SaCas9, the resulting mini-SaCas9 was nuclease-defective but retained efficient DNA binding activity. A set of compact transactivation domains (VTR1, VTR2, and VTR3) was then generated by removing the DNA binding domains in p65 and Rta. VTR1 was generated by substituting the p65 domain in the VPR with the TA1 and TA2 transactivation domains. VTR2 was created by replacing the p65 domain in the VPR with two repeats of the TA1. The partial Rta domain in the VTR1 was then removed to generate a VTR3. The resulting VTR1, VTR2 and VTR3 domains retain half of the transactivation efficiency of the full-length VPR.
In addition, self-assembled arrays of split SpyTag:SpyCatch or MoonTag:MoonCatcher peptides have been fused to the mini-SaCas9-VTR for recruiting multiple copies of VTR activator (Ma et al. 2018). SpyTag and MoonTag scaffold systems were smaller than the SunTag system for efficient packaging into the AAV vectors. To enable synergistic transcription activation using a single AAV expression cassette, self-cleaving ribozyme or small glutamine tRNA (∼70bp) has been used to replace the commonly used RNA polymerase III promoters (e.g. U6) (∼250bp) to simultaneous express multiple sgRNAs under the control of a single promoter (Xu et al. 2016). Ribozyme-linked sgRNAs enabled the use of tissue-specific RNA polymerase II promoters to express and release functional sgRNAs (He et al. 2017; Zhang et al. 2017). Meanwhile, tRNA-linked sgRNA fusion transcripts were efficiently and specifically cleaved by endogenous tRNase Z to release fully functional sgRNAs (Dong et al. 2017; Mefferd et al. 2015; Port and Bullock 2016). These tRNA- and ribozyme-linked sgRNA systems have been successfully implemented in the plant (He et al. 2017; Zhang et al. 2017) and mammalian cells (Dong et al. 2017; Xu et al. 2016). A recently developed single multiplex crRNA array can also be used to simultaneously target multiple sites in one gene (Sun et al. 2018). This single multiplex crRNA array utilized one U6 promoter to drive the expression of different direct repeats. Finally, truncated regulatory elements such as minimal promoters and terminators can be used to further reduce the transgene size for efficient AAV packaging.
To enable more robust epigenome editing and transcriptional modulation using AAV-CRISPR system, dCas9 conjugation through protein trans-splicing and dimerizing techniques can be adapted to recruit large endogenous chromatin regulators to specific genomic loci (Braun et al. 2017; Liszczak et al. 2017) (Figure 5). For example, Fkbp/Frb inducible recruitment for epigenome editing by dCas9 (FIRE-dCas9) system was a recently developed dCas9 conjugation approach that has been used to achieve rapid and reversible recruitment of endogenous chromatin regulators to specific genomic loci (Braun et al. 2017). Frb (FKBP-rapamycin-binding domain of mTOR) is a chemical-induced proximity tag that was fused to the desired chromatin regulator. Fkbp (FK506-binding-protein) is a complementary dimerizer of Frb that was fused to a dCas9-MS2 anchor. As a general principle, Fkbp/Frb dimerizing induced proximity of the Frb-chromatin regulator to dCas9-MS2-Fkbp upon rapamycin treatment. Their associations were reversed upon washout of the chemical dimerizer. This FIRE-dCas9 system has been successfully used to recruit endogenous Hp1/Suv39h1 heterochromatin and BAF chromatin-remodeling complexes to target genomic loci to modify local histone marks and rewire gene expression. For example, H3K9me3-mediated gene silencing was induced by recruiting Hp1/Suv39h1 heterochromatin complex to active promoters. By recruiting mSWI/SNF (BAF) complex to bivalent promoters, transcription activation was induced through the loss of repressive H3K27me3 and increase in active H3K4me3 marks. A chemically tailored dCas9 system was also recently developed by using protein trans-splicing to ligate synthetic elements (IntC-cargo such as IntC-JQ1 and IntC-UNC3866) to a dCas9-IntN (Liszczak et al. 2017). The resulting dCas9-IntN:IntC-cargo fusion was capable of recruiting endogenous copies of their cognate binding partners (e.g. Brd4T bromodomain and PRC1 chromodomain) to specific genomic loci. JQ1 is a small molecule bromodomain inhibitor that can interact with and recruit endogenous BET proteins (e.g. Brd4T bromodomain) by mimicking the acetyl-lysine mark. On the other hand, UNC3866 is a peptide-based PRC1 chromodomain ligand that can recruit and interact with the PRC1 complex.
Figure 5.
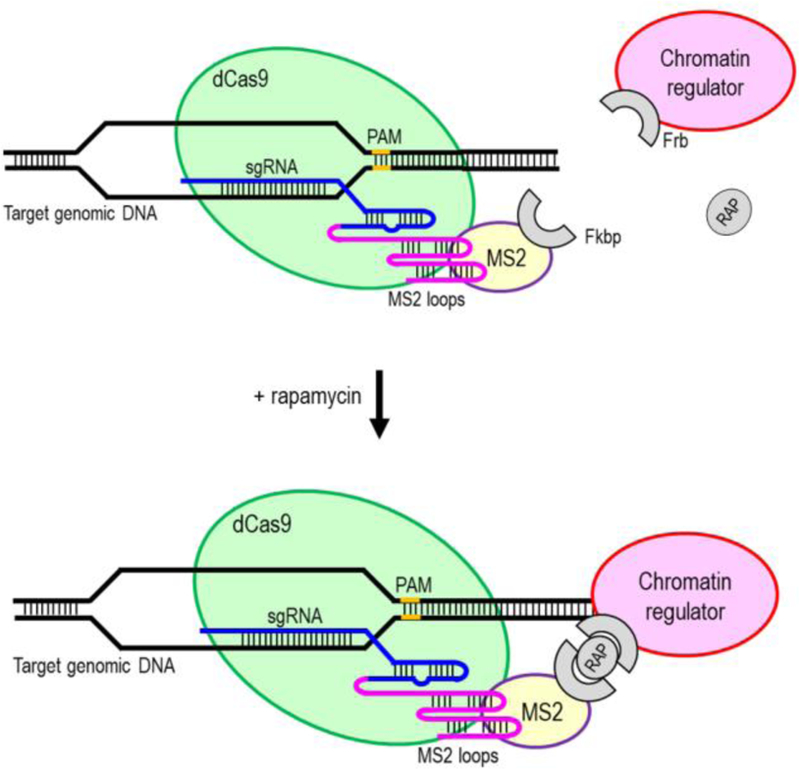
dCas9 conjugation approach to recruit endogenous chromatin regulators to specific genomic loci. Fkbp/Frb dimerizing induced proximity of the Frb-chromatin regulator to dCas9-MS2-Fkbp upon rapamycin (RAP) treatment.
Alternatively, a large catalytic domain was fused to the DNA binding domains of zinc finger nuclease (ZFN) and transcription activator-like effector nucleases (TALEN) for efficient packaging into the AAV vector. The ZFN (~1kb) and TALEN monomers (~3kb) are significantly smaller than the commonly used dCas9 (~4.2kb) (Gupta and Musunuru 2014). For example, the fusion of a TALE DNA binding domain to histone modifying enzyme such as H3K9me3 methyltransferase Suv39H1, H3K9me2 methyltransferase G9a (EHMT2) or the H3K9me2/3 demethylase JMJD2D (KDM4D) enabled modulation of H3K9 methylation at the target loci (Bieberstein et al. 2016). The TALE domain has also been fused to the catalytically active domain of SETD2 for increasing H3K36me3 levels at the target exon. In another study, TALE domain was fused to the LSD1 histone demethylase for downregulation of proximal genes by removing enhancer-associated chromatin modifications from target loci (Mendenhall et al. 2013). TALE domain has also been fused to the DNMT3A-Dnmt3L for stably silencing target gene expression by inducing DNA methylation and reducing chromatin accessibility at the promoters (Mlambo et al. 2018). In addition, the use of light-sensitive heterodimerizing proteins CRY2 and CIB1 enabled optogenetic control of epigenome editing. For example, dimerization of TALE-CIB1 with CRY2-DNMT3A or CRY2-TET1 enabled optogenetic induction of site-specific DNA methylation and demethylation, respectively (Lo et al. 2017). On the other hand, zinc finger-fused methyltransferases have been used for targeted DNA methylation of endogenous promoters and gene silencing (Kungulovski et al. 2015; Rivenbark et al. 2012). ZF-TET1 (Gallego-Bartolome et al. 2018) and TALE-TET1 (Maeder et al. 2013a) fusion proteins have also been successfully used for activation of endogenous genes by inducing targeted DNA demethylation at the methylated promoter CpG positions. Nevertheless, the use of ZFN and TALEN technologies are less popular than CRISPR owing to the difficulty and complexity in target designs and both TALENs and ZFNs require dimerization (Gaj et al. 2013).
Conclusion and perspective
CRISPR-based in vivo epigenome editing and transcriptional modulation have been achieved through the use of adeno-associated virus (AAV) delivery vehicle in adult mice, zygote microinjection, electroporation of embryos, generation of dCas9-expressing transgenic mice, and a genetic cross between dCas9- and sgRNA-expressing flies. Because AAV remains one of the most effective viral vectors to deliver CRISPR components in vivo, various AAV-compatible CRISPR systems have been developed for epigenome editing and transcriptional modulation in adult animals. These technological and conceptual advancements include the use of small dCas9 orthologues, truncated Cas9 mutants and transactivation domains, split-dCas9, and RNA-targeting systems. dCas9 conjugation has also helped tackle AAV packaging issue by recruiting endogenous chromatin modifiers and remodelers to specific genomic loci. Various protein scaffold systems such as SunTag, SAM, SpyTag, and MoonTag have also been developed to recruit multiple copies of dCas9 fusion proteins for robust epigenome editing and synergistic transcription activation or repression. Self-cleaving ribozyme and tRNA enable simultaneous expression of multiple sgRNAs under the control of a tissue-specific promoter. To simplify the AAV production and improve the delivery efficiency, various all-in-one AAV vector systems were developed by using truncated regulatory elements and a combination of the aforementioned strategies. The performance of the CRISPR-based epigenome editing in adult animals can be further improved by using the next-generation synthetic AAV capsids to minimize immunogenicity in vivo, and to enhance transduction efficiency and specificity in the tissues (Buning et al. 2015; Grimm and Buning 2017; Muzyczka and Warrington 2005).
Compared to genome editing, epigenome editing and transcriptional modulation offer several advantages in biomedical research and development of precision medicine or epigenetic therapies. Firstly, targeted epigenetic-oriented therapy can be used without the need to genetically modify the mutated DNA sequence or inducing DNA damage in order to correct gene misexpression, and to restore normal epigenetic patterning, chromatin structure, and nuclear organization. Thus, the potential creation of unwanted mutations within the target locus and off-target effects can be minimized. Secondly, many common human diseases are polygenic in origin (Yang et al. 2003) and associated with epigenome dysregulation (Hatchwell and Greally 2007). Given that a cis-regulatory element can interact and regulate multiple genomic loci (Jin et al. 2013; Mifsud et al. 2015; Sanyal et al. 2012), epigenetic therapy can provide a better therapeutic efficacy by simultaneous modulating multiple gene activities. Furthermore, the vast majority of genetic variants associated with the risk of common diseases mapped by genome-wide association studies (GWAS) are located in the non-coding regions of the genome and enriched with epigenomic marks (Kundaje et al. 2015). Epigenome editing can be complemented with genome editing to dissect the functional and mechanistic roles of these non-coding functional variants and associated genomic regulatory regions (Spisak et al. 2015). Thirdly, various catalytic domains of epigenetic effectors can be fused to dCas9 for studying the effects of a particular epigenetics regulation and gene-environment interaction. In addition to the DNA sequence, environmental factors can influence gene expression. For example, monozygotic twins may succumb to the same disease, but often the severity of symptom and age of onset are different (Fraga et al. 2005; Martin 2005). The phenotypic discordance of monozygotic twins, most likely due to epigenetic differences, became more noticeable with age. Fourthly, epigenome editing and transcriptional alteration are sufficient to induce pluripotency (Liu et al. 2018a), differentiation into a specific cell type or direct conversion between cell types (Black et al. 2016) without the need to use exogenous reprogramming factors or transgene integration into the host genome (Takahashi et al. 2007; Takahashi and Yamanaka 2006). Epigenome editing enabled rapid epigenetic remodeling and sustained transcriptional activation of endogenous master transcription factors. It is also reflected a more natural cell lineage conversion than forced overexpression of reprogramming factors, providing insights into the molecular mechanism of cellular reprogramming. Fifthly, epigenome editing allows stable (Amabile et al. 2016; Saunderson et al. 2017) or reversible (Braun et al. 2017; Nihongaki et al. 2017) manipulation of epigenetic states, depending on the types of catalytic domain or inducible strategy used. The edited epigenetic marks can be inherited throughout mitosis and somatic cell differentiation without the need for persistent modifier expressions (Amabile et al. 2016). Lastly, AAV delivery vehicles enable long-term expression of chromatin modifiers or transcriptional regulators in post-mitotic tissues of the adult animals without the need to integrate the transgenes into the host genome. Nevertheless, this field is still in its infancy and further refinements that increase the potency and heritability of epigenome editing are required.
Acknowledgements
This work was funded by NIH Grants (AG017242, GM104459, AG056278, and CA180126) and by the Glenn Center for the Biology of Human Aging (Paul Glenn Foundation for Medical Research) (Suh).
Footnotes
Conflict of interest
The author declares no conflict of interest.
References
- Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A (2016) Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167:219–232. https://doi:10.1016/j.cell.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V, Aigner S, Markmiller S, Xia G, Corbett KD, Swanson MS, Yeo GW (2017) Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170:899–912. https://doi:10.1016/j.cell.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Kozakova E, Huranova M, Thakur PK, Krchnakova Z, Krausova M, Carrillo Oesterreich F, Stanek D (2016) TALE-directed local modulation of H3K9 methylation shapes exon recognition. Sci Rep 6:29961 https://doi:10.1038/srep29961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JB, Adler AF, Wang HG, D’Ippolito AM, Hutchinson HA, Reddy TE, Pitt GS, Leong KW, Gersbach CA (2016) Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell 19:406–414. https://doi:10.1016/j.stem.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CJ, Bruno PM, Horlbeck MA, Gilbert LA, Weissman JS, Hemann MT (2016) Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc Natl Acad Sci USA 113:E3892–3900. https://doi:10.1073/pnas.1600582113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun SMG, Kirkland JG, Chory EJ, Husmann D, Calarco JP, Crabtree GR (2017) Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat Commun 8:560 https://doi:10.1038/s41467-017-00644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buning H, Huber A, Zhang L, Meumann N, Hacker U (2015) Engineering the AAV capsid to optimize vector-host-interactions. Curr Opin Pharmacol 24:94–104. https://doi:10.1016/j.coph.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Cano-Rodriguez D, Gjaltema RA, Jilderda LJ, Jellema P, Dokter-Fokkens J, Ruiters MH, Rots MG (2016) Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 7:12284 https://doi:10.1038/ncomms12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12:326–328. https://doi:10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491. https://doi:10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ding X, Feng Y, Seebeck T, Jiang Y, Davis GD (2017) Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun 8:14958 https://doi:10.1038/ncomms14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM (2016) A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 13:868–874. https://doi:10.1038/nmeth.3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. https://doi:10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- Colella P, Ronzitti G, Mingozzi F (2018) Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev 8:87–104. https://doi:10.1016/j.omtm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi:10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027. https://doi:10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Xie K, Chen Y, Yang Y, Mao Y (2017) Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem Biophys Res Commun 482:889–895. https://doi:10.1016/j.bbrc.2016.11.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugar G, Leenay RT, Eisenbart SK, Bischler T, Aul BU, Beisel CL, Sharma CM (2018) CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell 69:893–905. https://doi:10.1016/j.molcel.2018.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Yang-Zhou D, Fernandes VR, Gonzalez DP, Liu LP, Tao R, Ren X, Sun J, Hu Y, Zirin J, Mohr SE, Ni JQ, Perrimon N (2017) Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci USA 114:9409–9414. https://doi:10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12:1052–1058 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609. https://doi:10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF 3rd, (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405. https://doi:10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolome J, Gardiner J, Liu W, Papikian A, Ghoshal B, Kuo HY, Zhao JM, Segal DJ, Jacobsen SE (2018) Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA 115:E2125–E2134. https://doi:10.1073/pnas.1716945115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451. https://doi:10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Buning H (2017) Small but increasingly mighty: latest advances in AAV vector research, design, and evolution. Hum Gene Ther 28:1075–1086. https://doi:10.1089/hum.2017.172 [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145:423–434. https://doi:10.1016/j.cell.2011.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RM, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest 124:4154–4161. https://doi:10.1172/JCI72992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Shearwin KE, Dodd IB (2017) Programmable DNA looping using engineered bivalent dCas9 complexes. Nat Commun 8:1628 https://doi:10.1038/s41467-017-01873-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchwell E, Greally JM (2007) The potential role of epigenomic dysregulation in complex human disease. Trends Genet 23:588–595. https://doi:10.1016/j.tig.2007.08.010 [DOI] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, Kormaksson M, MacKerell AD Jr., Xue F, Mason CE, Hiebert SW, Prive GG, Cerchietti L, Bardwell VJ, Elemento O, Melnick A (2013) A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep 4:578–588. https://doi:10.1016/j.celrep.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang T, Yang N, Xu M, Yan L, Wang L, Wang R, Zhao Y (2017) Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J Genet Genomics 44:469–472. https://doi:10.1016/j.jgg.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517. https://doi:10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. https://doi:10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Su J, Lei Y, Brunetti L, Gundry MC, Zhang X, Jeong M, Li W, Goodell MA (2017) DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol 18:176 https://doi:10.1186/s13059-017-1306-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Xu RG, Ren X, Ewen-Campen B, Rajakumar R, Zirin J, Yang-Zhou D, Zhu R, Wang F, Mao D, Peng P, Qiao HH, Wang X, Liu LP, Xu B, Ji JY, Liu Q, Sun J, Perrimon N, Ni JQ (2018) Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc Natl Acad Sci USA https://doi:10.1073/pnas.1800677115 [DOI] [PMC free article] [PubMed]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B (2013) A high - resolution map of the three-dimensional chromatin interactome in human cells. Nature 503:290–294. https://doi:10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F (2017) Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. https://doi:10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12:401–403. https://doi:10.1038/nmeth.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8:14500 https://doi:10.1038/ncomms14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA (2017) CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol 35:561–568. https://doi:10.1038/nbt.3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K (2013) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32:267–273. https://doi:10.1038/nbt.2800 [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. https://doi:10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018) Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173:665–676. https://doi:10.1016/j.cell.2018.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. https://doi:10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kungulovski G, Nunna S, Thomas M, Zanger UM, Reinhardt R, Jeltsch A (2015) Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenetics Chromatin 8:12 https://doi:10.1186/s13072-015-0002-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DY, Zhao YT, Lamonica JM, Zhou Z (2017) Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat Commun 8:15315 https://doi:10.1038/ncomms15315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhang X, Su J, Jeong M, Gundry MC, Huang YH, Zhou Y, Li W, Goodell MA (2017) Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat Commun 8:16026 https://doi:10.1038/ncomms16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J, Li JF (2017) A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants 3:930–936. https://doi:10.1038/s41477-017-0046-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O’Keefe DD, Nunez-Delicado E, Guillen P, Campistol JM, Wu CJ, Lu LF, Esteban CR, Izpisua Belmonte JC (2017) In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171:1495–1507. https://doi:10.1016/j.cell.2017.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Ewen-Campen B, Ni X, Housden BE, Perrimon N (2015) In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics 201:433–442. https://doi:10.1534/genetics.115.181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszczak GP, Brown ZZ, Kim SH, Oslund RC, David Y, Muir TW (2017) Genomic targeting of epigenetic probes using a chemically tailored Cas9 system. Proc Natl Acad Sci USA 114:681–686. https://doi:10.1073/pnas.1615723114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Chen M, Liu Y, Qi LS, Ding S (2018a) CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell 22:252–261. https://doi:10.1016/j.stem.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R (2016) Editing DNA methylation in the mammalian genome. Cell 167:233–247. https://doi:10.1016/j.cell.2016.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, Li Y, Vershkov D, Cacace A, Young RA, Jaenisch R (2018b) Rescue of Fragile X Syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172:979–992. https://doi:10.1016/j.cell.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CL, Choudhury SR, Irudayaraj J, Zhou FC (2017) Epigenetic editing of Ascl1 gene in neural stem cells by optogenetics. Sci Rep 7:42047 https://doi:10.1038/srep42047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Peng S, Huang W, Cai Z, Xie Z (2018) Rational design of mini-Cas9 for transcriptional activation. ACS Synth Biol 7:978–985. https://doi:10.1021/acssynbio.7b00404 [DOI] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T (2015) Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA 112:3002–3007. https://doi:10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, Costello JF, Wilkinson MF, Joung JK (2013a) Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 31:1137–1142. https://doi:10.1038/nbt.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK (2013b) CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10:977–979. https://doi:10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. https://doi:10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM (2005) Epigenetic drift in aging identical twins. Proc Natl Acad Sci USA 102:10413–10414. https://doi:10.1073/pnas.0504743102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefferd AL, Kornepati AV, Bogerd HP, Kennedy EM, Cullen BR (2015) Expression of CRISPR/Cas single guide RNAs using small tRNA promoters. RNA 21:1683–1689. https://doi:10.1261/rna.051631.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE (2013) Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 31:1133–1136. https://doi:10.1038/nbt.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, Herman B, Happe S, Higgs A, LeProust E, Follows GA, Fraser P, Luscombe NM, Osborne CS (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47:598–606. https://doi:10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]
- Mingozzi F, High KA (2011) Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12:341–355. https://doi:10.1038/nrg2988 [DOI] [PubMed] [Google Scholar]
- Mlambo T, Nitsch S, Hildenbeutel M, Romito M, Muller M, Bossen C, Diederichs S, Cornu TI, Cathomen T, Mussolino C (2018) Designer epigenome modifiers enable robust and sustained gene silencing in clinically relevant human cells. Nucleic Acids Res 46:4456–4468. https://doi:10.1093/nar/gky171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AM, Fu X, Zhu J, Katrekar D, Shih YV, Marlett J, Cabotaje J, Tat J, Naughton J, Lisowski L, Varghese S, Zhang K, Mali P (2018) In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol Ther 26:1818–1827. https://doi:10.1016/j.ymthe.2018.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, Hoffman AR, Huang CC, Pitteri SJ, Wang KC (2017) Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8:15993 https://doi:10.1038/ncomms15993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, Hatada I (2016) Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34:1060–1065. https://doi:10.1038/nbt.3658 [DOI] [PubMed] [Google Scholar]
- Muzyczka N, Warrington KH Jr., (2005) Custom adeno-associated virus capsids: the next generation of recombinant vectors with novel tropism. Hum Gene Ther 16:408–416. https://doi:10.1089/hum.2005.16.408 [DOI] [PubMed] [Google Scholar]
- Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M (2017) CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods 14:963–966. https://doi:10.1038/nmeth.4430 [DOI] [PubMed] [Google Scholar]
- O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, Mackay JP, Farnham PJ, Segal DJ (2017) dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res 45:9901–9916. https://doi:10.1093/nar/gkx578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M (2016) Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533:125–129. https://doi:10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA (2013) RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10:973–976. https://doi:10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Bullock SL (2016) Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods 13:852–854. https://doi:10.1038/nmeth.3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D, Raya A (2017) CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21:431–447. https://doi:10.1016/j.stem.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. https://doi:10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, Flynn EM, Romero FA, Magnuson S, Gascoigne KE (2018) Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep 24:1722–1729. https://doi:10.1016/j.celrep.2018.07.041 [DOI] [PubMed] [Google Scholar]
- Ran F, Cong L, Yan W, DA S, Gootenberg J, Kriz A, Zetsche B, Shalem O, Wu X, Makarova K, Koonin E, Sharp P, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191. https://doi:10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P (2012) Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7:350–360. https://doi:10.4161/epi.19507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu MJ, Bloom JS, Day L, Siegel JJ, Kosuri S, Kruglyak L (2018) Highly parallel genome variant engineering with CRISPR-Cas9. Nat Genet 50:510–514. https://doi:10.1038/s41588-018-0087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489:109–113. https://doi:10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunderson EA, Stepper P, Gomm JJ, Hoa L, Morgan A, Allen MD, Jones JL, Gribben JG, Jurkowski TP, Ficz G (2017) Hit-and-run epigenetic editing prevents senescence entry in primary breast cells from healthy donors. Nat Commun 8:1450 https://doi:10.1038/s41467-017-01078-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senis E, Fatouros C, Grosse S, Wiedtke E, Niopek D, Mueller AK, Borner K, Grimm D (2014) CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J 9:1402–1412. https://doi:10.1002/biot.201400046 [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. https://doi:10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, Ye H (2018) Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc Natl Acad Sci USA 115:E6722–E6730. https://doi:10.1073/pnas.1802448115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. https://doi:10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Spisak S, Lawrenson K, Fu Y, Csabai I, Cottman RT, Seo JH, Haiman C, Han Y, Lenci R, Li Q, Tisza V, Szallasi Z, Herbert ZT, Chabot M, Pomerantz M, Solymosi N, Consortium G-OE, Gayther SA, Joung JK, Freedman ML (2015) CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat Med 21:1357–1363. https://doi:10.1038/nm.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, Reik W, Jeltsch A, Jurkowski TP (2017) Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45:1703–1713. https://doi:10.1093/nar/gkw1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover JD, Farhang N, Berrett KC, Gertz J, Lawrence B, Bowles RD (2017) CRISPR epigenome editing of AKAP150 in DRG neurons abolishes degenerative IVD-induced neuronal activation. Mol Ther 25:2014–2027. https://doi:10.1016/j.ymthe.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA (2018) RNA-dependent RNA targeting by CRISPR-Cas9. Elife 7 https://doi:10.7554/eLife.32724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li F, Liu J, Yang F, Zeng Z, Lv X, Tu M, Liu Y, Ge X, Liu C, Zhao J, Zhang Z, Qu J, Song Z, Gu F (2018) A single multiplex crRNA array for FnCpf1-mediated human genome editing. Mol Ther 26:2070–2076. https://doi:10.1016/j.ymthe.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F (2015) In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33:102–106. https://doi:10.1038/nbt.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi:10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. https://doi:10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, Gersbach CA (2016) Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 13:127–137. https://doi:10.1038/nmeth.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA (2015) Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12:1143–1149. https://doi:10.1038/nmeth.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Kwon JB, Nelson CE, Rouse DC, Gemberling MP, Oliver ML, Gersbach CA (2018) RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun 9:1674 https://doi:10.1038/s41467-018-04048-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellinga J, Smith JP, Lipiec A, Majhen D, Lemckert A, van Ooij M, Ives P, Yallop C, Custers J, Havenga M (2014) Challenges in manufacturing adenoviral vectors for global vaccine product deployment. Hum Gene Ther 25:318–327. https://doi:10.1089/hum.2014.007 [DOI] [PubMed] [Google Scholar]
- Voets O, Tielen F, Elstak E, Benschop J, Grimbergen M, Stallen J, Janssen R, van Marle A, Essrich C (2017) Highly efficient gene inactivation by adenoviral CRISPR/Cas9 in human primary cells. PLoS One 12:e0182974 https://doi:10.1371/journal.pone.0182974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V (2016) Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44:5615–5628. https://doi:10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora S, Cheng J, Xiao R, VanDusen N, Quintino L, Pu W, Vandenberghe L, Chavez A, Church G (2018) Rational design of a compact CRISPR-Cas9 activator for AAV-mediated delivery. bioRxiv https://doi.org/10.1101/298620
- Wangensteen KJ, Wang YJ, Dou Z, Wang AW, Mosleh-Shirazi E, Horlbeck MA, Gilbert LA, Weissman JS, Berger SL, Kaestner KH (2017) Combinatorial genetics in liver repopulation and carcinogenesis with a novel in vivo CRISPR activation platform. Hepatology 68:663–676. https://doi:10.1002/hep.29626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltner J, Balboa D, Katayama S, Bespalov M, Krjutskov K, Jouhilahti EM, Trokovic R, Kere J, Otonkoski T (2018) Human pluripotent reprogramming with CRISPR activators. Nat Commun 9:2643 https://doi:10.1038/s41467-018-05067-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Senanayake U, Artibani M, Taylor G, Wells D, Ahmed AA, Sauka-Spengler T (2018) Genome and epigenome engineering CRISPR toolkit for in vivo modulation of cis-regulatory interactions and gene expression in the chicken embryo. Development 145 https://doi:10.1242/dev.160333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA (2016) Biology and applications of CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell 164:29–44. https://doi:10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- Wu H, Mathioudakis N, Diagouraga B, Dong A, Dombrovski L, Baudat F, Cusack S, de Massy B, Kadlec J (2013) Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep 5:13–20. https://doi:10.1016/j.celrep.2013.08.035 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao L, Gao Y, Xu J, Han R (2016) Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res 45:e28 https://doi:10.1093/nar/gkw1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA (2018) Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell 70:327–339. https://doi:10.1016/j.molcel.2018.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Botto L, Friedman JM, Flanders WD (2003) Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Genet 72:636–649. https://doi:10.1086/367923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM (2016) A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 34:334–338. https://doi:10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo NC, Chavez A, Lance-Byrne A, Chan Y, Menn D, Milanova D, Kuo CC, Guo X, Sharma S, Tung A, Cecchi RJ, Tuttle M, Pradhan S, Lim ET, Davidsohn N, Ebrahimkhani MR, Collins JJ, Lewis NE, Kiani S, Church GM (2018) An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat Methods 15:611–616. https://doi:10.1038/s41592-018-0048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Gao Y, Wang R, Zhao Y (2017) Production of guide RNAs in vitro and in vivo for CRISPR using ribozymes and RNA polymerase II promoters. Bio Protoc 7 https://doi:10.21769/BioProtoc.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Shen W, Zhang J, Yang B, Liu YN, Qi H, Yu X, Lu SY, Chen Y, Xu YZ, Li Y, Gage FH, Mi S, Yao J (2018) CRISPR interference-based specific and efficient gene inactivation in the brain. Nat Neurosci 21:447–454. https://doi:10.1038/s41593-018-0077-5 [DOI] [PubMed] [Google Scholar]
- Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, Yang H (2018) In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci 21:440–446. https://doi:10.1038/s41593-017-0060-6 [DOI] [PubMed] [Google Scholar]


