Figure 10.
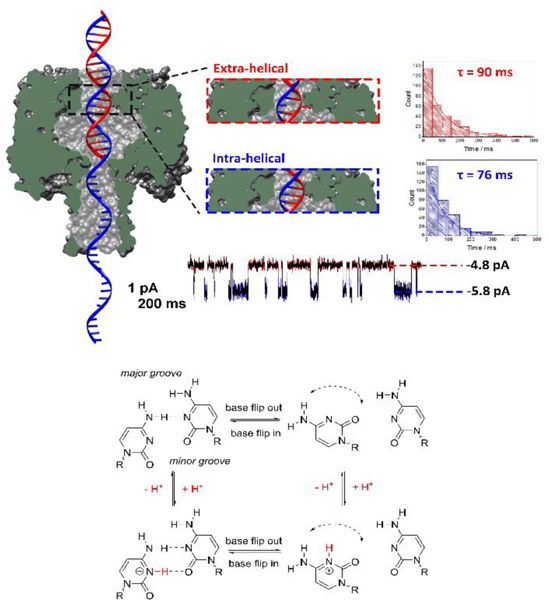
Measuring the dynamics of the motion of an individual DNA base by monitoring the ionic current through a protein nanochannel. (Top) Cross-section of the α-HL protein nanopore with a double-stranded DNA molecule (with a single-stranded tail) captured in the vestibule. The dsDNA contains a cytosine-cytosine mismatch near the latch zone of the protein. The observed current displays a two state modulation on the millisecond timescale resulting from the dynamic base flipping of one of the cytosine bases between intrahelical and extrahelical states. Histograms of the duration times between the two current states, and single exponential fits, are used to measure the first-order lifetimes of the extra-helical (out of the helix) and intra-helical (within the helix) states. (Bottom) Square scheme depicting the protonation/deprotonation and the effect of pH on the number of hydrogen bonds within cytosine-cytosine base pair. Below the pKa of the cytosine mismatch, an additional hydrogen bond is formed decreasing the rate of base flipping. Although base flipping is a relatively slow process, the protonation/deprotonation that affects the flipping kinetics is immeasurably fast. Figure adapted from reference 99.
